Advances in Animal and Veterinary Sciences
Research Article
Identification of some Domestic Animal Species (Camel, Buffalo and Sheep) by PCR-RFLP Analysis of the Mitochondrial Cytochrome b Gene
Mayada Ragab Farag1*, Tamer Said Imam1, Kuldeep Dhama2
1Department of Forensic Medicine and Toxicology, Faculty of Veterinary Medicine, Zagazig University, Zagazig 44111, Egypt; 2Division of Pathology, Indian Veterinary Research Institute, Izatnagar, Bareilly, 243122, Uttar Pradesh, India.
Abstract | The present study describes the use of PCR and PCR-RFLP methods to discriminate between blood samples obtained from domestic camel, buffalo and sheep. The DNA was extracted from 3 blood samples of each species at 3 different ages and subjected to PCR amplification of a mitochondrial cytochrome b (cyt b) gene segment (358 bp) using universal primers. Subsequent cleavage with 4 restriction endonucleases (AluI, HaeIII, HinfI and Taq I) gaverise to a species-specific pattern on an agarose gel. Results of cleavage by AluI and HaeIII were 2 fragments of different sizes in all animals; Hinf I produced 2 fragments in camel and sheep and 3 fragments in buffalo; Taq I produced 2 bands in camel and sheep while no fragmentation was generated for buffalo. The results suggest that the method of PCR-RFLP using these restriction enzymes can reliably identify chosen species. Moreover, PCR-RFLP is a rapid and simple method for identification of animal species.
Keywords | Species identification, Animals, Cytochrome b gene, PCR-RFLP, Forensic samples
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | January 06, 2015; Revised | January 24, 2015; Accepted | January 25, 2015; Published | January 30, 2015
*Correspondence | Mayada Ragab Farag, Zagazig University, Zagazig, Egypt; Email: [email protected]
Citation | Farag MR, Imam TS, Dhama K (2015). Identification of some domestic animal species (camel, buffalo and sheep) by PCR-RFLP analysis of the mitochondrial cytochrome b gene. Adv. Anim. Vet. Sci. 3(2): 136-142.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.2.136.142
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Farag et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Determining the species origin of different kind of samples (blood, meat, skin, etc.) is important in forensic purpose for species differentiation and identification (Sherryn et al., 2015) and it is also an integral part of food regulatory issue as adulteration and substitution of meat has always been a concern for various reasons such as public health, religious factors, wholesomeness and unhealthy competition in meat market (Rashid et al., 2014; Hou et al., 2015).Various methods like traditional morphological or protein identification methods, species specific markers or repeats (SSR), mitochondrial DNA restricted fragment length polymorphism (mtDNA-RFLP), and random amplified polymorphic PCR (RAPD-PCR) are employed for detection of the species origin of different forensic samples and meat, and among these DNA based assays are gaining popularity (Cheng et al., 2014; Lin et al., 2014). Particularly the mitochondrial DNA (mtDNA) has been the most widely studied region of eukaryotic genomes which has played a critical role in development of population and evolutionary genetics (Abou-Hadeed et al., 2011; DeMasi et al., 2015). It has several advantages over nuclear genome for diagnostic studies of animals because of greater abundance in sample extracts and higher copy number making the amplification of mtDNA segment to be a relatively sensitive procedure (Sumathi et al., 2015).
The cytochrome b gene (cyt b) encodes one of the best known proteins that make up complex III of the mitochondrial phosphorylation system and is the only one encoded by the mitochondrial genome. The cyt b gene is used as an important utility tool in studies of legal medicine and molecular evolution (Prusak et al., 2004; Abou-Hadeed et al., 2011). This gene has been completely sequenced or partially sequencedfor many species of mammals, birds (Bravi et al., 2004; Andrzej and Kamila, 2005), reptiles, amphibian and fishes (Chow et al., 1993; Ram et al., 1996; Quinterio et al., 1998; Lidstrom, 1999; Parson et al., 2000) and also some invertebrates (Lee et al., 2009a).
Species identification studies based on the cyt b gene have ranged from both short (< 400 base pair) to long (> 900 base pair) PCR-RFLP DNA sequencing and variable size species specific multiplex PCR (Lee et al., 2009b). An alternative DNA detection system is based on the PCR amplification of a segment of the mitochondrial cyt b gene followed by RFLP analysis based on subsequent cleavage by a restriction enzymes which gives rise to species specific pattern (Ahmed, 2007; Abd El-Rahman et al., 2009). This technique requires less equipment and cost than sequencing.
One of the most important issues in forensic medicine is to identify the sample’s origin and discriminate whether they descend from human or non-human origins. Therefore, the present study was aimed atexploit the molecular technique of PCR-RFLP analysis of the mitochondrial cytochrome b gene followed by subsequent cleave using 4 restriction enzymes (AluI, HaeIII, HinfI and Taq I) for differentiation between camel, buffalo and sheep. These restriction enzymes don’t used before for identification of camel and do not used collectively in these species where HinfI and Taq I have been used only for buffalo not for camel and sheep identification.
MATERIALS AND METHODS
Samples
The 5 ml blood samples were collected from domestic camel, buffalo and sheep (3 each from each different age: new born, young and adult), reared in local animal farms in Zagazig, Egypt. The DNA was extracted from the blood samples of camel, buffalo and sheep using QIAamp® tissue kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. The concentration and purity of the extracted DNA samples were measured by NanoDrop® ND-1000 (Full- Spectrum UV Spectrophotometer at 260/280 nm) as presented in Table 1.
The blood DNA samples of the three animal species were subjected to PCR amplification of the mitochondrial cytochrome b gene. The primers employed were: Primer 1 (L 14816) (5´- CCA TCC AAC ATC TCA GCA TGATGA AA – 3’) = P1: conc. 0.25 µM; Primer 2 (H15173) (5’-CCC CTC AGA ATG ATA TTT GTC CTC A-3’)= P2: conc. 0.25 µM (Metabion international AG, Deutschland).The PCR Master mix (RED TaqReadyMix PCR Reaction Mix, with MgCl2) used comprised of a total volume of 25 µl containing 20 mMTris- HCl, Ph 8.3, with 100MKCl, 3 mM MgCl2, 0.002% gelatin, 0.4 mMdNTP mix (dATP, dCTP, dGTP, TTP), stabilizers, and 0.06 unit/µl of Taq DNA polymerase (Sigma) and nuclease free water (NFW, PCR reaction water) (Sigma).The amount of PCR mix, primers, DNA samples and water used in the PCR reaction for amplification of the cyt b gene fragment from the blood samples of the three animal species is presented in Table 2.
Table 1: The concentration and purity of DNA extracted from blood samples
|
The sample |
Concentration (ng/ µl) |
Purity (at 260/280nm) |
|
Camel |
40.20 |
1.8 |
|
Buffalo |
90.77 |
1.8 |
|
Sheep |
190.3 |
1.4 |
Table 2: PCR reaction - PCR mix, primers, DNA sample and water used in amplification of cyt b gene fragment of three animal species
|
PCR requirements |
Camel |
Buffalo |
Sheep |
|
RED Taqreadymix (µl) |
12.5 |
12.5 |
12.5 |
|
Forward primer (µl) |
1 |
1 |
1 |
|
Reverse primer (µl) |
1 |
1 |
1 |
|
DNA template (sample) (µl) |
2.5 |
3.5 |
4 |
|
PCR reaction water (µl) |
8.0 |
7.0 |
6.5 |
|
Total volume (µl) |
25 |
25 |
25 |
Table 3: RFLP banding pattern of cyt b gene PCR amplified fragment afterdigestion with different restriction endonucleases
|
Animal species |
Restriction endonuclease cleavage pattern |
|||
|
Alu I |
HaeIII |
HinfI |
Taq I |
|
|
Camel |
2 (198+160) |
2 (238+120) |
2 (300+58) |
2 (185+173) |
|
buffalo |
2 (290+68) |
2 (259+99) |
3(195+145+48) |
1 (358) |
|
Sheep |
2 (312+46) |
2 (278+80) |
2(200+158) |
2 (260+98) |
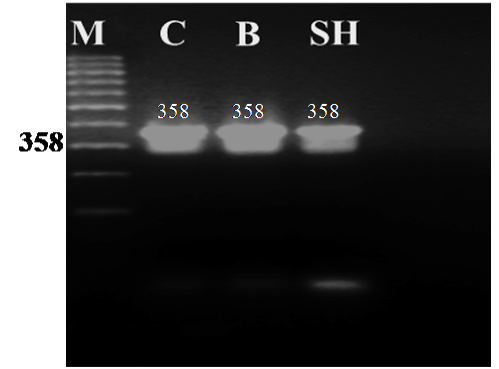
Figure 1: Agarose gel electrophoresis of PCR amplified products (358 bp) of the cytochrome b gene from blood samples of camel (C), buffalo (B) and sheep (SH)
The PCR protocol as described by Bartlett and Davidson (1992) was employed for amplification of the cytochrome b gene with the extracted DNA samples. A 5µl of the extracted DNA sample was used. The PCR amplification reaction conditions comprised of a total volume of 25µl consisting of 1X PCR reaction buffer (20 mMTris- HCl, pH 8.3, with 100 mMKCl, 3 mM MgCl2), 0.4 mMdNTP, 0.25µM each primer and 0.06 unit/µl of Taq DNA polymerase (Perkin Elmer), DNA templates ( 2.5, 3.5 and 4.0µl for camel, buffalo and sheep samples, respectively). The PCR cyclic conditions comprised of 35 cycles at 94°C for 30 seconds, 50°C for 45 seconds and 72°C for 45 seconds including one initial denaturation step at 95°C for 11 minutes in a 9600 GeneAmp (Perkin Elmer) thermocycler. The PCR amplified samples were electrophoresed on 1.6% agarose gel in 0.5 X TBE buffer at 100-150 Ma for 30-45 minutes. The visualization of the DNA fragments was done in a UV cabinet unit and photographed with a Polaroid camera.
PCR-RFLP Analysis
For RFLP analysis, the PCR amplified products (10 ul) were directly digested with 5 U of each one of the four restriction endonuclease enzymes viz., AluI, HaeIII, Hinf I and Taq I separately (Roche Diagnostics, Roche applied science, Mannheim, Germany). The digestion was performed for 2 hours at 37°C and the digested DNA products were separated by electrophoresis on 3% agarose gels, and visualized under UV illumination and photographed with a Polaroid camera. The length of the restriction fragments generated after RE digestion was compared with the standard 100 bp DNA Ladder (100 to 1000 bp scale) (Jena Bioscience, GmbH, Germany).
RESULTS
The universal primers of L14816 and H15173 were used to amplify part of the cyt b gene (358 bp) from blood samples of camel, buffalo and sheep. The PCR generated a single amplification product for each genomic DNA template from the three animal species. The size of all the PCR products from the different animals tested showed no obvious differences when separated on 1.6 % agarose gel with the expected amplicon size (358 bp) of the cyt b gene as shown in Figure 1.
The different size of bands produced under gel electrophoresis after digestion with the four restriction endonucleases (AluI, HaeIII, Hinf I and Taq I) were identified by comparison of the standard size marker using graphical methods. The cleavage patterns of the four RE enzymes of the cyt b gene PCR amplified products of the three animal species including their band numbers and fragment sizes are presented in Table 3. The RFLP analysis of the cyt b gene region of the three different animal species revealed high specificity and discrimination, even there was no common fragment shared among the studied species. Moreover the resulting fragments were the same for each species in all samples regardless the age (newborn, young and adult) .Restriction endonucleases digestion of the PCR products showed specific patterns as follows: Alu I yielded 2 restriction fragments of different sizes in camel, buffalo and sheep (Figure 2A); HaeIII also revealed 2 restriction fragments of different sizes patterns in all species (Figure 2B); Hinf I produced 2 fragments of different sizes for both camel and sheep and 3 fragments in buffalo (Figure 2C); Taq I produced 2 bands differing in their sizes between camel and sheep while no fragmentation was generated for buffalo by this enzyme (Figure 2D).The resulted fragments by the 4 different restriction enzymes in all species are completely identical to those obtained using hair and meat samples from the same species (data not shown).
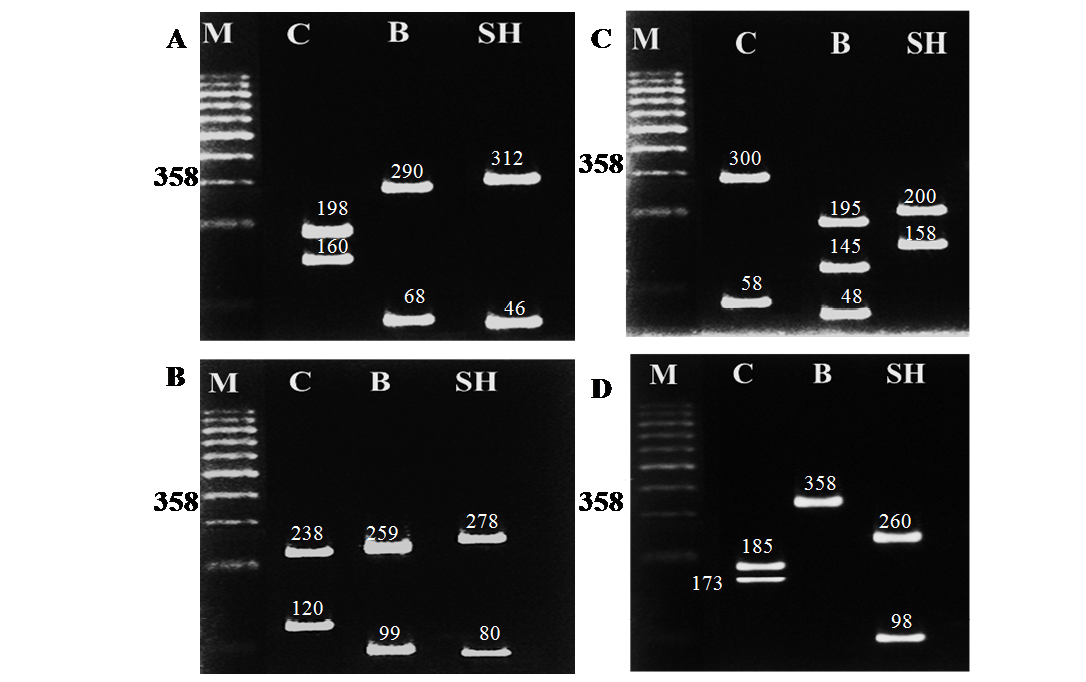
Figure 2: RFLP banding pattern of the cyt b gene PCR amplified products after digestion with different restriction endonucleases. A: AluI, B: HaeIII, C: Hinf I, D: Taq I
DISCUSSION
DNA identification methods generally give better resolution and confirmatory identification than the traditional morphological or protein identification methods, and are the most useful tools for determining animal species in commercial foods and animal products (Ali et al., 2015; Andrea et al., 2015). When analysis of such animal samples is under taken for species level identification it usually involves sequencing of all or part of the mitochondrial genome which is then compared to known sequences in Gene Bank (NCBI). This technique is accepted, but has many problems including: being costly, time and labour intensive due to the extra step of sequencing products, mixtures cannot be separated, and the generated samples may not produce sufficient sequence data (Lenstra et al., 2001). The more modern techniques now allow the identification of species specific markers or repeats (SSR), mitochondrial DNA restricted fragment length polymorphism (mtDNA-RFLP), and random amplified polymorphic PCR (RAPD-PCR) which have the major advantages over protein analysis and that the samples heated to as high as 120°C can still be analysed and discriminated between species (Minarovič et al., 2010; Linacre, 2012; Muhammad and Yasmeen, 2015).
In the present study, a short DNA fragment (358 bp) of the mitochondrial cytochrome b gene was investigated and exploited for species identification of three domestic animals (camel, buffalo and sheep) because this fragment has by far the widest taxonomic representation in nucleotide databases as reported by Guo et al. (2005). The PCR amplification of the cyt b gene, using universal primers (L14816 and H15173) generated a specific 358 bp product with DNA samples of all the three animal species tested, which is in accordance to the report of (Parson et al., 2000).The resulting PCR amplified products when digested with four restriction endonucleases (AluI, HaeIII, Hinf I and Taq I) revealed different RFLP patterns which discriminated the samples with animal species origin. In theoretical case of the same fragments of chosen species we recommend to pick different restriction enzyme for RFLP identification.
The selection of these enzymes for further experiments was based on their putative ability which discriminate between the samples of different animal species in polyacrylamide gels. In theoretical case of the same fragments of chosen species we recommend to pick different restriction enzyme for RFLP identification.
The RFLP analysis of the cyt b gene region with the samples of the different animal species (camel, buffalo and sheep) showed high specificity and discriminatory power that even there was no common digestion fragment shared among them. Restriction endonucleases digestion showed specific patterns for each animal species, and these results were confirmatory to that obtained by Apostolidis et al. (2000) who used AluI, HaeIII, Hinf I and Taq I to screen five horse breeds in Greek and found these enzymes to have restriction sites as recognition sites on cyt b gene and were able to identify polymorphism and classify the horse breeds studied. The results of the present study also are in agreement and support of the results obtained by Bravi et al. (2004) who reported that the PCR- RFLP method allowed them to identify cattle, horse, donkey, pig, sheep, dog, cat, rabbit, chicken, and human through the different restriction patterns of the enzymes AluI, HaeIII, Hinf I on cyt b gene fragment (358 bp). They concluded that this method may be useful as a first assessment process in forensic evidence and testing for species level identification.
The results obtained by AluI enzyme were supported by Akasaki et al. (2006) who found that AluI enzyme generated different digestion patterns enabling identification of cod fish imported into Japan and consequently they found that the PCR-RFLP methods were sufficient for rapidly screening the cod products. These results were also confirmed by those of Minarovič et al. (2010) who reported that Alu I enzyme generated different digestion patterns enabling identification of Mustelavison (81 bp, 109 bp and 169 bp), Mustelaputoriusfuro (169 bp and 190 bp), Susscrofadomesticus (115 bp and 244 bp), Oryctolaguscunninculus is not cleaved by AluIso it has whole 359 bp fragment on agarose gel, Anseranser (130 bp and 229 bp). Abd El-Rahman et al. (2009) have also used species specific primers to identify cat’s, dog’s, donkey’s and horse’s meat, they could differentiate between cat’s, dog’s, and horse’s meat but the same PCR amplification size of horse (221 bp) was obtained for donkey and to differentiate between the two species they used restriction enzyme AluI which yielded three fragments in horse’s meat (189, 96 and 74 bp), whereas no fragments were obtained in donkey’s meat.
Regarding to the results obtained by HaeIII enzyme, similar results were reported by Viseshakul and Sarikaputi, (2003) in their study to identify Bosgaurus origin from the DNA recovered from other possible ruminant species by PCR-RFLP of cyt b gene using HaeIII, and stated that this method could give conclusive results. Similarly, Jung et al. (2008) concluded that the PCR-RFLP was proved to be a rapid, reliable and simple method that enabled the identification of six commercial filefish processed species, where they amplified a cyt b gene fragment (465 bp) from processed filefish meats. Then they exposed the PCR amplified fragment to digestion by HaeIII enzyme which could differentiate the species of A. monoceros (285+180 bp), A. scriptus (180+159+126 bp), M. chinensis (424+41 bp), T. hypergyrus (180+159+94+32 bp), T. modestus (229+180+56 bp) and C. penicilligerus (285+139+41bp).
PCR-RFLP of cyt b gene has also been used by Nagata et al. (2005) to differentiate Tiger (Pantheratigris) and Leopard (Pantherapardus) using HinfI enzyme which appeared more likely to produce easily distinguishable differences in RFLP banding patterns between the two wild animal species and these results are in agreement with the results of the present study concerning using HinfI enzyme for identification of animal species.
Our investigation concerning the use of Taq I enzyme was proved by previous study carried out by Ahmed et al. (2007) in which they amplified a segment of cyt b gene (359 bp) and exposed it to digestion by TaqI restriction enzyme to discriminate between buffalo’s and cattle’s meat, which generated two fragments (191 and 168 bp) in buffalo whereas the cyt b gene fragment was not digested in cattle because using of the species specific primers couldn’t differentiate between the two species. The results showed that the cytochrome b gene PCR-RFLP analysis provides rapid and effective method to detect species origin of given samples. The results obtained by the four enzymes used in the present study are totally in agreement with those of recent study of Abou-Hadeed et al. (2011) where these enzymes could successfully differentiate between hair samples of American black bear (Urusamericanus), blue nile monkey (Ceropithecusmitis), Barbary sheep (Amotracuslervia), bacterian camel (Camelusbactrianus), llama (Lama glama) and human (Homo sapiens).
CONCLUSION
This study shows that species identification method based on PCR- amplification of a segment of the mitochondrial cytochrome b gene, processed with RFLP analysis using the four restriction endonucleases (AluI, HaeIII, HinfI and Taq I) can be very reliable. It can also be considered as a simple, quick and cheap method in forensic identification of the studied animal species (camel, buffalo and sheep). Application of genetic methods is very useful for breeding of livestock and protection of biodiversity as well as differentiation of sample´s origin which could be valuable in detection of adulteration.
REFERENCES





