Advances in Animal and Veterinary Sciences
Research Article
Molecular Characterization of Canine Parvovirus-2 (Cpv-2) in Dogs in Egypt
Rabie Etman*, Mahmoud S. Safwat, H. Khodeir
Faculty of Veterinary Medicine, Cairo University, Egypt.
Abstract | Canine parvovirus-2 (CPV-2) causes a severe, often fatal, enteric disease in dogs worldwide. Molecular characterization of CPV-2 should be regularly carried out because of its high evolution rate. The present study aimed to molecularly characterize CPV-2 field strains, circulating in Giza Governorate, Egypt. A total of 22 rectal swabs collected from dogs with clinical signs suggestive of canine parvovirus enteritis were initially screened for CPV-2 using field (CPV-2 Ag rapid kit) and molecular (conventional PCR, using the primer pairs ParvoInt2FB/ParvoInt2CR) techniques. All swabs were further examined by conventional PCR, using the sequencing primer pairs 555for/555rev; samples that had showed strong PCR products were sequenced. Results showed all swabs (n = 22) were positive for CPV-2 by conventional PCR, using the primer pairs ParvoInt2FB/ParvoInt2CR, whereas 20 samples were positive for CPV-2 Ag rapid kit. All samples were positive for conventional PCR, using the primer pairs 555for/555rev; only 16 samples were further sequenced and molecularly characterized as CPV-2a (12/16, 75%), CPV-2c (3/16, 18.75) and CPV-2b (1/16, 6.25%). Additionally, Thr440Ala mutation was prevalent in the characterized samples (13/16, 81.25%). The majority of samples (16/22, 72.2%) were obtained from non-vaccinated dogs. Therefore, it could be concluded that there is cross-immunity between commercial vaccine strains (original strain or CPV-2b) and CPV-2a, the predominant variant in the study area; however, further challenge studies are necessary to confirm this conclusion.
Keywords | CPV-2; Egypt; Molecular characterization; Rapid kit.
Received | October 27, 2020; Accepted | November 13, 2020; Published | May 25, 2021
*Correspondence | Rabie Etman, Faculty of Veterinary Medicine, Cairo University, Egypt; Email: Rabieetman.vetcu@gmail.com
Citation | Etman R, Safwat MS, Khodeir H (2021). Molecular characterization of canine parvovirus-2 (cpv-2) in dogs in egypt. Adv. Anim. Vet. Sci. 9(6): 933-940.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.6.933.940
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Etman. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Canine parvovirus type 2 (CPV-2) is a small, non-enveloped DNA virus protected by a capsid, which is responsible for the antigenic properties, host range, tissue tropism and hemagglutination activity of the virus (Tsao et al., 1991).
CPV-2 suddenly emerged in 1978, most likely as a variant of another parvovirus hosted by wild red foxes (Truyen et al., 1998). CPV-2 evolved into three antigenic variants termed CPV-2a, CPV-2b and CPV-2c during the years 1979, 1984 and 2001, respectively (Parrish et al., 1988; Parrish et al., 1991; Buonavoglia et al., 2001).
The question of whether there is a cross-protection between these variants is still under debate (Decaro et al., 2020). These variants differ in only one amino acid (aa) residue at position 426 of the major capsid protein (VP2) as follows: asparagine (Asn) in CPV-2a, aspartic acid (Asp) in CPV-2b and glutamic acid (Glu) in CPV-2c (Parrish et al., 1991, Buonavoglia et al., 2001).
According to the age of affected dogs, CPV-2 causes either non-suppurative myocarditis or enteritis; canine parvovirus enteritis is the most commonly reported form and clinically characterized by sudden onset of fever, anorexia, depression, foul-smelling diarrhea, often hemorrhagic, vomiting and dehydration in non-vaccinated dogs, mostly 2-6-month-old puppies (Decaro and Buonavoglia, 2012).
For the field diagnosis of CPV-2, a variety of commercial fecal CPV-2 Ag rapid kits can be used in the veterinary clinics, whereas for the laboratory diagnosis, viral isolation, hemagglutination test, and conventional and real-time PCR are the most commonly used techniques (Mylonakis et al., 2016). CPV-2 could be effectively controlled by the using of modified live virus (MLV) vaccines and proper disinfectants, such as sodium hypochlorite (Decaro and Buonavoglia, 2012).
CPV-2 has a high substitution rate for its genome, resembling that of the highly evolving RNA viruses; what is more, a few mutations in the VP2 could result in major antigenic variations (Decaro and Buonavoglia, 2012).
The existence of new CPV-2 variants and mutants could impact both the performance of diagnostic techniques, especially those depending on monoclonal antibodies, and the efficacy of vaccines (Truyen, 2006; Kapil et al., 2007; Decaro et al., 2009; Decaro et al., 2010; Demeter et al., 2010; Decaro and Buonavoglia, 2012; Amrani et al., 2016; Decaro et al., 2020a). Therefore, the periodical performance by DNA sequencing of well-designed CPV-2 molecular characterization studies is epidemiologically necessary (Decaro and Buonavoglia, 2012).
Taking into account the lack of such studies in Egypt despite their abovementioned epidemiological importance, this work was performed to molecularly characterize CPV-2 field strains, circulating in the Giza Governorate, Egypt from May to August 2019.
Materials and Methods
Sampling
All procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee of the Faculty of Veterinary Medicine, Cairo University (Vet CU01102020227). A total of 22 rectal swabs were collected from dogs with clinical signs suggestive of canine parvovirus enteritis. These dogs were admitted to the teaching hospital of the Faculty of Veterinary Medicine, Cairo University, Egypt, in the period from May to August 2019. Rectal swabs were mixed with 2 ml (10% w\v) phosphate-buffered saline solution (PH 7.2) then transported on ice to the laboratory, where these samples were vigorously mixed, using a vortex, for 30 sec then centrifuged at 1500 x g for 10 minutes. The supernatants were kept frozen at – 20 °C till use.
Field and laboratory diagnosis of CPV-2
For the field diagnosis, animal’s signalment and history were recorded, physical examination of the affected dogs was performed, and rectal swabs were tested in the hospital for the presence of CPV-2 antigens, using the VDRG® CPV Ag Rapid kit (Median diagnostic co, Korea), following the manufacturer’s instructions.
For the laboratory diagnosis, viral DNA was extracted from all rectal swabs, using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany), following the manufacturer’s protocol. All samples were screened for the presence of CPV-2 DNA by conventional PCR, using the screening primer pairs ParvoInt2FB/ ParvoInt2CR (Faz et al., 2017), which partially amplify the VP2 gene. All data regarding these primer pairs (sequences, polarity and positions) are summarized in Table 1, whereas the thermal conditions of PCR reactions are given in Table 2. The positive result is indicated by the visualization of PCR products of 275 bp length.
All PCR reactions were conducted in the Applied Biosystem Thermal Cycler (Biometra, Germany). A commercial monovalent CPV-2 MLV vaccine “primodog” (Merial, France) and nuclease-free PCR grade water (Thermo Fischer Scientific, USA) were used as a positive and negative control, respectively. PCR products were separated by 1.5% agarose gel electrophoresis and the gel was photographed by a gel documentation system (Biometra, Germany).
CPV-2 antigenic variants typing
All samples were further examined for CPV-2 by conventional PCR, using the sequencing primer pairs 555for/555rev (Buonavoglia et al., 2001); samples that had showed strong PCR products were subjected to sequencing. The primer pairs 555for/555rev amplify a part of the VP-2 gene, encoding the aa residue at position 426. All data regarding these primer pairs (sequences, polarity and positions) are summarized in Table 1, whereas, the thermal conditions of PCR reactions are given in Table 2. The positive result is marked by the visualization of amplicons of 583 bp length.
The PCR products were purified from the gel by the QIAquick Gel Extraction Kit (Qiagen, Germany). The samples were sequenced by the Sanger method in a forward direction, using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Thermo Fisher Scientific, USA).
The obtained sequences were translated to the corresponding aa sequences, using the BioEdit sequence alignment editor software version 7.2 (Hall, 1999). The deduced aa sequences were aligned with four references CPV-2 aa sequences retrieved from GenBank [the original strain (M38245.1), CPV-2a (M24003.1), CPV-2b (M74849.1), and CPV-2c (FJ222821.1)], using the clustal W multiple alignment option of the BioEdit software.
Table 1: The use, sequence, polarity and position of the primer pairs ParvoInt2FB/ParvoInt2CR and 555for/555rev
| Name | Use | Sequence (polarity) | Position c |
| ParvoInt2FB | Screening | TCAAGCAGCAGATGGTGATCCAAG (+) | 3892-3915 |
|
ParvoInt2CR a |
GGTACATTATTTAATGCAGTTA (-) | 4167–4146 | |
|
555for |
Sequencing | CAGGAAGATATCCAGAAGGA (+) | 4003–4022 |
|
555rev b |
GGTGCTAGTTGATATGTAATAAACA (-) |
4585–4561 |
a Reference; Faz et al. (2017)
b Reference; Buonavoglia et al. (2001)
c Oligonucleotide positions are referred to the genomic sequence of CPV-2 strain CPV-b (GenBank accession no. M38245).
Table 2: The thermal conditions of conventional PCR technique, using the primer pairs ParvoInt2FB/ParvoInt2CR and 555for/555rev
| The primer pairs | Thermal conditions | |||||
| Initial denaturation | Amplification | Final extension | ||||
| Number of cycles | Denaturation | Annealing | extension | |||
|
ParvoInt2FB/ ParvoInt2CR a |
94 ◦C for 5 min |
35 |
94 ◦C for 30 s |
55◦C for 1 min |
72 ◦C for 1 min |
72 ◦C for 5 min |
|
555for/ 555rev b |
94 ◦C for 10 min |
40 |
94 ◦C for 30 s |
50 ◦C for 1 min |
72 ◦C for 1 min |
72 ◦C for 10 min |
a Reference; Faz et al. (2017) b Reference; Desario et al. (2005)
Results
Field and laboratory diagnosis
All data regarding the collected animal’s signalment and vaccination history are summarized in Table 3. The owners complain and the physical examination revealed that out of 22 examined dogs, 3 dogs (13.6%) showed fever, anorexia, depression, bloody, foul-smelling diarrhea, bloody vomiting and dehydration; 5 dogs (22.75%) suffered from anorexia, depression, bloody, foul-smelling diarrhea and depression; 5 dogs (22.75) had anorexia, non-bloody diarrhea and dehydration; 5 dogs (22.75%) displayed anorexia, bloody, foul-smelling diarrhea and dehydration, and 4 dogs (18.1%) displayed anorexia and depression.
Concerning the results of the fecal CPV-2 Ag rapid kits, 20 samples were positive for CPV-2 antigens (20/22, 91%). Regarding the results of the laboratory diagnosis, all rectal swabs were positive for conventional PCR, using the primer pairs ParvoInt2FB/ParvoInt2CR, i.e., amplicons were 275 bp in length, as seen in Figure 1.
CPV-2 antigenic variant Typing
All samples were positive for conventional PCR, using the sequencing primer pairs 555for/555rev, i.e., amplicons were 583 bp in length, as shown in Figure 2.
Sixteen samples that had showed strong PCR bands were sequenced; all of these samples were successfully sequenced and the obtained sequences (n = 16) were deposited in GenBank with the following accession numbers:
MT636872- MT636874 and MT596684-MT596696.
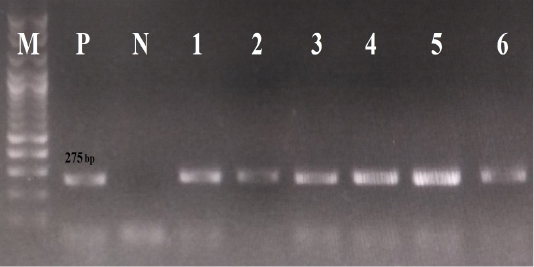
Figure 1: The results of conventional PCR technique, using the primer pairs ParvoInt2FB/ ParvoInt2CR: the positive reaction indicated by an amplicon that is 275 bp in length (M: DNA ladder 100bp, P: positive control (Primodog, Merial, France), N: negative control (PCR grade water), 1-6: positive samples obtained in this study).
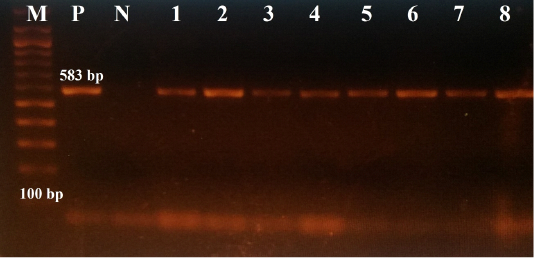
Figure 2: The results of conventional PCR technique, using the primer pairs 555for/555rev: the positive reaction indicated by an amplicon that is 583 bp in size (M: DNA ladder 100bp, P: positive control (Primodog, Merial, France), N: negative control (PCR grade water), 1-8: positive samples obtained in this study).
Table 3: The signalment and vaccination history of studied animals, and CPV-2 antigenic variants characterized in this study.
| ID | Animal signalment | Vaccination history | CPV-2 antigenic variant (Accession Number) | |||
| Age month | Sex | Breed | Vaccination status (Vaccine strain) | Time since last vaccination | ||
| Vetcu-1 | 6 | ♂ | German shepherd | Non-vaccinated | - |
b (MT636874) |
| Vetcu-2 | 2 | ♂ | Rottweiler |
Vaccinated (CPV-2b) |
1 week |
a (MT596684) |
| Vetcu-3 | 11 | ♂ | Labrador Retrieve | Non-vaccinated | - |
c (MT596685) |
| Vetcu-4 | 5 | ♂ | German shepherd | Non-vaccinated | - |
a (MT596686) |
| Vetcu-5 | 4 | ♂ | Native | Non-vaccinated | - |
c (MT596687) |
| Vetcu-6 | 2 | ♂ | German shepherd | Non-vaccinated |
-
|
a (MT596688) |
| Vetcu-7 | 3.5 | ♀ | Golden Retriever | Non-vaccinated | - |
a (MT596689) |
| Vetcu-8 | 2 | ♂ | Griffon | Non-vaccinated | - |
a (MT596690) |
| Vetcu-9 | 2.5 | ♀ | Cocker Spaniel |
Vaccinated (CPV-2b) |
2 weeks |
a (MT596691) |
| Vetcu-10 | 3 | ♀ | Stray | Non-vaccinated | - |
a (MT596692) |
| Vetcu-11 | 4 | ♂ | Golden Retriever |
Vaccinated (CPV-2b) |
3 weeks |
a (MT596693) |
| Vetcu-12 | 7 | ♂ | Griffon | Non-vaccinated | - |
a (MT596694) |
|
Vetcu-13 |
6 | ♀ | Rottweiler | Non-vaccinated | - |
a (MT596695) |
| Vetcu-14 | 3 | ♀ | Beagle |
Vaccinated (CPV-2) |
1.5 months |
c (MT636872) |
| Vetcu-15 | 7 | ♀ | German shepherd | Vaccinated (CPV-2) | 4 months |
a (MT596696) |
| Vetcu-16 | 5 | ♀ | Rottweiler | Non-vaccinated | - |
a (MT636873) |
| Vetcu-17 | 2.5 | ♂ | Golden Retriever | Non-vaccinated | - |
-
|
|
Vetcu-18 |
6 |
♂ |
Siberian Husky |
Vaccinated (CPV-2) |
3 months |
- |
|
Vetcu-19 |
1.5 | ♀ |
Saint Bernard |
Non-vaccinated |
- | - |
|
Vetcu-20 |
2.5 | ♀ |
Rottweiler |
Non-vaccinated | - | - |
|
Vetcu-21 |
2.5 | ♀ |
Rottweiler |
Non-vaccinated | - | - |
|
Vetcu-22 |
2 |
♂ |
Caucasian |
Non-vaccinated | - | - |
As shown in Figure 3 and Table 3, 75% (12/16), 18.75% (3/16) and 6.25% (1/16) of samples were molecularly characterized as CPV-2a, CPV-2c and CPV-2b, respectively; in addition, the mutation from thyronine (Thr) to alanine (Ala) at position 440 (Thr440Ala) was prevalent in the characterized samples (13/16, 81.25%).
Discussion
The lack of well-designed studies, discussing CPV-2 evolution in Egypt, encourages us to perform the present study to molecularly characterize CPV-2 field strains, circulating in the Giza Governorate, Egypt in the period from May
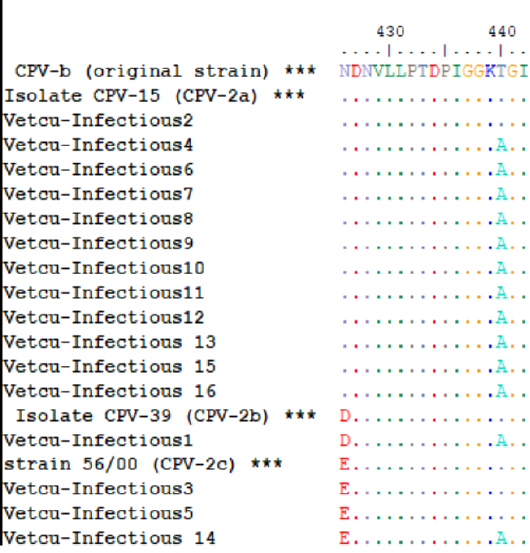
Figure 3: The multiple amino acid alignments of the Egyptian sequences with reference strains for CPV-2 antigenic typing; the Egyptian sequences (Vet Cu-Infectious 1-16, accession numbers: MT596684-MT596696 and MT636872-MT636874) were aligned together with reference sequences (asterisks) that include [CPV-b (original strain, M38245.1), CPV-15 (CPV-2a, M24003.1), CPV-39 (CPV-2b, M74849.1) and strain 56/00 (CPV-2c, FJ222821.1)], which indicates that CPV-2a was the predominant variant.
to August 2019.
In the present study, at the time of their presentation to veterinary clinics, only 3 out of 22 affected dogs (3/22, 13.6%) showed all of the clinical signs described for canine parvovirus enteritis in previous literature (fever, anorexia, depression, foul-smelling, bloody diarrhea, vomiting and dehydration) (Decaro and Buonavoglia, 2012), whereas other dogs displayed variable combinations of some of these clinical signs; among them, four dogs showed only anorexia and depression.
Similarly, such variability has been frequently reported (Kalli et al., 2010; Aldaz et al., 2013; Faz et al., 2017) and it might occur due to several factors, such as the time to presentation, infectious dose of virus, affected animal’s age and immune status, and whether is there a mixed infection (Faz et al., 2017).
The diagnostic implication of such variability is the false-negative results for the clinical diagnosis of CPV-2 infected puppies because most of the veterinary practitioners would clinically suspect canine parvovirus enteritis only when puppies are admitted with foul-smelling, bloody diarrhea (Faz et al., 2017; M. Safwat, personal observation).
All dogs were examined for the presence of CPV-2 in their rectal swabs, using the in-clinic fecal CPV-2 Ag rapid kits and conventional PCR, using the primer pairs ParvoInt2FB/ParvoIn2CR. The conventional PCR technique has a high analytical sensitivity and subsequently, it could detect the presence of even a low virus titer (Faz et al., 2017). Conversely, due to its low analytical sensitivity, the fecal CPV-2 Ag rapid kits might not detect the presence of this low virus titer, resulting in false-negative results (Desario et al., 2005; Decaro and Buonavoglia, 2012).
In the current work, two dogs displayed false-negative results with the rapid kit, possibly due to the shedding of a low virus titer in the stool. The first dog was admitted at a late stage of the illness (fourth day) when CPV-2 specific antibodies are expected to be present (Decaro and Buonavoglia, 2012). These specific antibodies neutralize the virus, reducing its titer in the stool (Desario et al., 2005; Decaro and Buonavoglia, 2012; Decaro et al., 2013). The other dog displayed severe, frequent, large volume watery diarrhea that could reduce the virus titer in the stool by its dilutional effect (Mylonakis et al., 2016).
In the present study, 16 samples were molecularly characterized by sequencing. Because MLV vaccine strains are expected to be shed in the stool for four weeks after its administration (Friesl et al., 2017), CPV-2 molecular characterization has a diagnostic value, discriminating between CPV-2 field and vaccine strains in dogs, showing gastroenteritis shortly after CPV-2 MLV vaccines administration (Decaro and Buonavoglia, 2012).
The deduced aa sequences obtained in this study have only one informative aa residue at position 426 of the VP2, which is the only aa residue that differ between the three antigenic variants (Asn in CPV-2a, Asp in CPV-2b and Glu in CPV-2). This aa residue is identical (426Asn) in both the original strain and CPV-2a (Parrish et al., 1991); the original strain is extinct in the field and it presents only in commercial vaccines (Parrish et al., 1991; Gamoh et al., 2003; Desario et al., 2005). Therefore, this aa residue could not differentiate between CPV-2a and the original strain only in dogs recently vaccinated with the original strain-based vaccines or with unknown vaccination history (Castro et al., 2011).
Because all recently vaccinated dogs in this study have been vaccinated with CPV-2b-based vaccines, as given in table 3, and their obtained sequences have 426 Asn, it could be concluded that all recently vaccinated dogs were infected with field rather than vaccine strains.
When it is performed periodically, CPV-2 molecular characterization by sequencing has also an epidemiological importance because it could rapidly and early detect the existence of CPV-2 new antigenic variants and mutants, which is expected to be frequently occur due to the high evolution rate of CPV-2 (Decaro and Buonavoglia, 2012).
There is a debate in the literature regarding the role of the evolution of such antigenic variants and mutants as a risk factor for CPV-2 vaccination failure. For example, Decaro et al. (2009) reported a case of CPV-2 vaccination failure in an adult dog has been annually vaccinated against CPV-2, possibly due to the antigenic differences between the vaccine strain (original strain) and the detected field strain (CPV-2c). In contrast, Schultz et al. (2010) proved, through a challenge study, that the original strain-based vaccine could protect against CPV-2c for 5 years.
CPV-2 variations could also result in the misdiagnosis of CPV-2 by the commonly used field test (CPV-2 Ag rapid kits); for example, after the emergence of CPV-2c, Kapil et al. (2007) raised concerns regarding the ability of the commercially available fecal CPV-2 Ag rapid kits to detect this new variant; however, in a study conducted by Decaro et al. (2010), the authors found that CPV-2c could be detected by the SNAP Canine Parvo Antigen Test.
Additionally, mischaracterization of CPV-2 by methods other than sequencing have been reported due to the existence of such mutants; for instance, Demeter et al. (2010) noted by sequencing that a mutation, which created a new restriction site for the restriction enzyme MobII at the 5-prime end of the VP-2 in all of the Hungarian CPV-2a strains, has resulted in the mischaracterization by the MobII-based restriction fragment length polymorphism (RFLP) technique of all of the examined samples as CPV-2c.
The data generated in this study indicate that CPV-2a (426Asn) was the predominant CPV-2 antigenic variant and 440Ala was the prevalent mutant, circulating in the Giza governorate, Egypt, in the period from May to August 2019. These aa residues are in a major antigenic site termed A, which is located on the top of the threefold spikes (Tsao et al., 1991). In the present study, it was noted that firstly, the majority of the studied samples were obtained from non-vaccinated (n = 16) rather than vaccinated dogs (n = 6)—secondly, most of the examined samples gave true-positive results with the rapid kits. These findings suggest that the existence of this antigenic variant or mutant in the Egyptian CPV-2 strains might not impact the performance of the field diagnostic test nor the efficacy of the commonly used vaccines in the study area.
There are variable patterns of the geographical distribution among different CPV-2 antigenic variants and mutants (Miranda and Thompson, 2016). The reason and epidemiological significance for these differences are not well-understood. However, it may be caused by migration events (Maya et al., 2013, Mira et al., 2018), non-representative sampling (Decaro et al., 2011) or local adaptations; Thr440Ser mutation, for instance, is found only in the Ecuadorian and Peruvian CPV-2 isolates (Aldaz et al., 2013; De la Torre et al., 2018; Quispe et al. (2018). Similarly, the mutations Phe267Tyr, Tyr324Ile and Gln370Arg are commonly used as a marker for the Asiatic CPV-2 strains (Mira et al., 2018).
According to the results of the latest molecular surveillance studies of CPV-2 from other countries, CPV-2a was the prevalent variant in South Korea in 2007 (Yoon et al., 2009), Greece from 2008 to 2009 (Ntafis et al., 2010), New Zealand from 2009-2010 (Ohneiser et al., 2015), Hungary in 2012 (Csagola et al., 2014), Albania from 2011 to 2013 (Cavalli et al., 2014), Pakistan from 2014 to 2015 (Ahmed et al., 2018), India from 2014 to 2015 (Kulkarni et al., 2019), Lithuania from 2014 to 2015 (Zienius et al., 2016), The Caribbean island of St. Kitts from 2015-2016 (Navarro et al., 2017), Poland from 2008 to 2016 (Kowalczyk et al., 2019), China from 2016 to 2017 (Zhuang et al., 2019), Ecuador in 2017 (De la Torre et al., 2018) and Colombia from 2018 to 2019 (Giraldo-Ramirez et al., 2020).
Thr440Ala mutant was also found in a large number of countries, for examples, South Korea in 2007 (Yoon et al., 2009), South Africa and Mozambique during the year 2010 (Dogonyaro et al., 2013; Figueiredo et al., 2017), Uruguay from 2009-2011 (Maya et al., 2013), Pakistan from 2014 to 2015 (Ahmed et al., 2018), Mexico in 2016 (Faz et al., 2019), Zambia in 2017 (Kapiya et al., 2019), Chile from 2016-2017 (Castillo et al., 2020), China from 2016 to 2017 (Zhuang et al., 2019), Brazil from 2015-2018 (de Oliveira et al., 2019), Nigeria in 2018 (Ogbu et al., 2020) and Colombia from 2018 to 2019 (Giraldo-Ramirez et al., 2020).
The limitations of this study are the small number of samples and the short-sequenced part of the VP-2, which contains little informative aa residues. Therefore, another study that involves a larger number of samples with full VP-2 gene sequencing should be performed for a better understanding of CPV-2 evolution in Egypt.
Conclusions
The majority of samples were taken from non-vaccinated (16/22); therefore, it could be concluded that the predominant antigenic variant, CPV-2a, and mutant 440Ala, might not impact the successful immunization by the commercial vaccines that are available in the study area (the original strain and CPV-2b-based vaccines). However, further well-designed challenge studies are required to confirm this conclusion.
Conflict of interest
The authors declare that there is no conflict of interest.
References






