Advances in Animal and Veterinary Sciences
Research Article
Rapid Detection of Enterotoxigenic Staphylococcus aureus Isolated from Raw Cow Milk in Sharkia Governorate, Egypt
Sally H. Abou-Khadra1*, Ibrahim M. El-Shorbagy2, Mona M. El-Azzouny1
1Microbiology Unit, Animal Health Research Institute(AHRI), Zagazig branch, Agriculture Research Center (ARC), Egypt; 2Food Hygiene Unit, Animal Health Research Institute(AHRI), Dokii, Giza, Agriculture Research Center (ARC), Egypt.
Abstract | Staphylococcus aureus (S. aureus) produces various types of toxins, that cause food poisoning. This study was designed for a rapid investigation of enterotoxigenic S. aureus from raw milk. One hundred raw milk samples were collected from different farms in Sharkia governorate and examined for genome corresponding to enterotoxin genes (Sea, Seb, Sec, Sed, and See) of S. aureus using multiplex polymerase chain reaction (M-PCR). A reverse passive latex agglutination assay (RPLA) was also used for screening the enterotoxin production. The presence of S. aureus was detected in 20% of the examined samples. The antimicrobial susceptibility patterns showed sensitivity to ciprofloxacin (100%), followed by vancomycin (96%). Maximum resistance was reported for oxacillin (100%), ampicillin (95%) and amoxicillin (90%). Staphylococcal enterotoxin D gene (Sed) was detected in all isolates, while Seb and Sec genes were not detected. Though the results of PCR and RPLA had congruence, a few discrepancies were also reported for some of the isolates. The study concludes the detection of enterotoxigenic S. aureus (Sed and Sea gene) in raw milk samples, and therefore, ascertain necessary interventions to avoid food poisoning.
Keywords | Staphylococcal enterotoxins, Multiplex polymerase chain reaction, Reversed passive latex agglutination, Raw milk
Received | June 10, 2020; Accepted | June 25, 2020; Published | July 12, 2020
*Correspondence | .Sally H. Abou-Khadra; Microbiology Unit, Animal Health Research Institute(AHRI), Zagazig branch, Agriculture Research Center (ARC), Egypt; Email: [email protected], sallylab85@gmail
Citation | Abou-Khadra SH, El-Shorbagy IM, El-Azzouny MM (2020). Rapid detection of enterotoxigenic Staphylococcus aureus isolated from raw cow milk in Sharkia governorate, Egypt. Adv. Anim. Vet. Sci. 8(s1): 11-17.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.s1.11.17
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Abou-Khadra et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Staphylococcus aureus (S. aureus) has emerged as one of the most important pathogens over the past several decades. It is considered a leading cause of food-poisoning outbreaks and contagious bovine mastitis (Guven et al., 2010; El-Jakee et al., 2013). Milk is regarded as a suitable medium for its growth, where potential contamination occurs during processing stages (Asao et al., 2003; Le Loir et al., 2003; Brasca et al., 2005; Jørgensen et al., 2005).
The leading cause of raw milk contamination with S. aureus is the dairy cows. These are mainly the cows that suffer from subclinical mastitis, and therefore had the potential to disseminate this microbe intomilk (Da Silva et al., 2005). The success of this pathogen for producing toxin is ascribed to its potential virulence, where, with the emergence of multi-drug resistance (MDR), it has proven remarkable ability to overcome most of the antibiotics developed in the recent years (Diep et al., 2008). Infection caused by antibiotic resistant strains such as methicillin-resistant S. aureus (MRSA) is considered severe threat to the healthcare system worldwide, mainly due to ease in its spread and the difficulties inpatient treatment. This is primarily due to the dissemination of determinants that encode resistance to antimicrobial drugs (Witte, 1999). There were five classes of SE types were previously identified (SEA, SEB, SEC, SED and SEE) (Bergdoll, 1983).
In the 1990s, sequence analyses discovered further “new” Se genes These are Sek, Sel, Sem, Sen, Seo, Sep, Seq, Ser, and Seu; however, their importance in food poisoning has not been explicated (Morandi et al., 2007; Boynukara et al., 2008; Rall et al., 2008).
Staphylococcal enterotoxins (SEs) proteins are about 26.900 - 29.600 Kilo Dalton, These are resistant to heat and the proteolytic enzymes in the gastrointestinal tract (Martın et al., 2004). These are not affected by pasteurization, For instance, Staphylococcal enterotoxin A (SEA) keeps some activity even after exposure to heat at 121°C for 28 min (Rall et al., 2008). A consumption of staphylococcal enterotoxin SEA is the most frequent cause of food poisoning (Stewart, 2003), and several outbreaks are attributed accordingly (Asao et al., 2003). It is the leading cause of staphylococcal food poisoning outbreaks in the developed countries where it represents about 75% of outbreaks followed by SED, SEB, and SEC (Normanno et al., 2005).
Multiplex PCR is required for the simultaneous identification of S. aureus enterotoxin genes. However, gene existence does not confirm the enterotoxigenic properties of a strain. Therefore, an expression of the gene also should be measured (Van Belkum, 2003).
Recently enzyme-linked immunosorbent assays were employed for direct detection of SE in food. However, these methods are time consuming and do involve a high cost. On the other hand, reverse passive latex agglutination (SET-RPLA) is considered the best method for the identification of SE in bacterial culture (Morandi et al., 2007) that can detect enterotoxin protein using specific antibodies (Sergeev et al., 2004). The present work aimed to identify enterotoxin genes Ses (Sea, Seb, Sec, Sed and See) among multidrug-resistant S. aureus strains using multiplex polymerase chain reaction (PCR), and SEs protein production using the RPLA technique.
Material and Methods
Sampling
During the period from March to October 2018, one hundred raw milk samples were randomly collected from farms in Sharkia Governorates, Egypt. Milk samples were incubated at 37°C for 24 h, centrifuged at 3000 rpm for 20 minutes, and the sediment was used for bacteriological cultivation.
Bacteriological examination
All samples were subjected to the routine preliminary phenotypic characterization of S. aureus isolates on the basis of standard microbiological techniques (Baird,1996).
Antimicrobial susceptibility testing
In vitro antimicrobial susceptibility testing of the S. aureus isolates against a panel of 9 antimicrobial agents was tested by the Kirby-Bauer standard agar disk diffusion technique as described earlier (Bauer, 1966). The tested antibiotics and their concentrations in μg/disc included clindamycin (DA; 2 μg), tetracycline (TE; 30), ciprofloxacin (CIP; 5μg), oxacillin (OX; 1μg), vancomycin (VA; 30 μg), amoxicillin/clavulanic acid (AMC; 20/10 μg), amoxicillin (Ax; 10 μg), ampicillin (AM; 10 μg) and gentamycin (CN; 10 μg) (Oxoid Ltd., Basingstoke, Hampshire, UK). Inhibition zone, (mm) was measured in duplicate and scored in accordance with the critical breakpoints recommended by the Clinical and Laboratory Standards Institute (CLSI, 2014). All S. aureus colonies were screened for methicillin resistance using oxacillin.
Multiplex PCR for the detection of enterotoxin genes
A total of ten S. aureus isolates were used for genome extraction that showed the highest multidrug-resistant. DNA extraction was performed using the QIAamp DNA Mini kit (Qiagen, Germany, GmbH). Briefly, 200 µl of the sample suspension was incubated with 10 µl of proteinase K and 200 µl of lysis buffer at 56oC for 10 min. After incubation, 200 µl of 100% ethanol was added to the lysate. The sample was then washed and centrifuged following the manufacturer’s recommendations. Nucleic acid was eluted with 100 µl of elution buffer. The primer sequences, their corresponding genes and the respective molecular sizes of PCR amplified products are listed in Table 1. PCR amplification reactions were performed in a final volume of 25 μL containing 12.5 μL of DreamTaq TM Green Master Mix (2X) (Fermentas, Inc. Hanover, USA), 0.1 μL of 100 pmol of each primer (SigmaAldrich, Co., St. Louis, USA), and 2 μL of S. aureus DNA template. The volume of the reaction mixture was completed to 25 μL using DNase/RNase-free water. The cycling condition was carried out using a PTC-100 TM programmable thermal cycler (MJ Research Inc., Waltham, USA). PCR was performed with an initial denaturation at 94˚C /5 min, followed by 35 cycles of each of 94°C for 30 sec, 57°C for 40 sec, and 72°C for 40 sec, and a final extension of 10 min at 72°C (Mehrotra et al., 2000).
PCR product analysis
The PCR products were separated by electrophoresis on 1.5% agarose gel (Applichem, Germany, GmbH) in 1x TBE (Tris-borate-EDTA) buffer at room temperature using gradients of 5V/cm. The fragment sizes of PCR products were determined with a 100 bp DNA ladder (Qiagen, Germany), photographed, and the data was analyzed through computer software.
Staphylococcal enterotoxin (SE) detection by the RPLA method
The detection of enterotoxin production (SEA, SEB, SEC, SED) from broth cultures of S. aureus isolates were detected by commercially available kit (SET-RPLA,TD 900, Oxoid, UK) according to manufacturer’s instructions.
Table 1: Primers sequences, target genes, amplicon sizes and cycling conditions.
| Target gene | Primers sequences | Amplicon size (bp) |
| Sea | GGTTATCAATGTGCGGGTGG | 102 |
| CGGCACTTTTTTCTCTTCGG | ||
| Seb | GTATGGTGGTGTAACTGAGC | 164 |
| CCAAATAGTGACGAGTTAGG | ||
| Sec | AGATGAAGTAGTTGATGTGTATGG | 451 |
| CACACTTTTAGAATCAACCG | ||
| Sed | CCAATAATAGGAGAAAATAAAAG | 278 |
| ATTGGTATTTTTTTTCGTTC | ||
| See | AGGTTTTTTCACAGGTCATCC | 209 |
| CTTTTTTTTCTTCGGTCAATC |
Statistical analysis
Data analysis was performed using the statistical package for the Social Sciences (SPSS), version 22. The chi-square test was used at a 5% significance level.
Results
Occurrence and distribution of S. aureus among the examined samples
Out of 100 raw milk samples, 30 of them (30%) were positive for Staphylococcus spp. However, the coagulase test illustrated only 20 isolates (20 %) were coagulase positive (S. aureus) and, therefore, considered as pathogenic strains.
Antimicrobial susceptibility testing
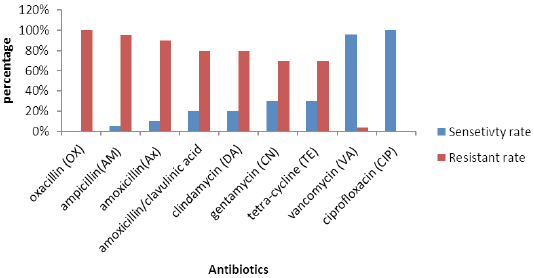
Antimicrobial susceptibility patterns of all coagulase positive (S. aureus) isolates are illustrated in Figure 1. Analysis of methicillin resistance confirmed that all the recovered isolates were MRSA. All tested MRSA isolates were susceptible to ciprofloxacin (100%) and vancomycin (96%). However, higher frequency levels of resistance were obtained for ampicillin (95%), followed by amoxicillin (90%) clindamycin (80%). All the isolates were considered resistant to three or more of the antimicrobial agents of three different groups and therefore taken as multiple drug resistance (MDR).
S. aureus enterotoxigenic genes
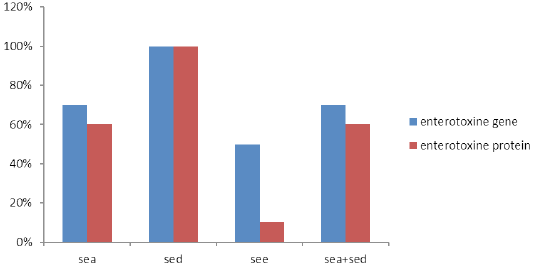
S. aureus isolates from raw milk samples were examined by multiplex PCR, where ten isolates showed their genomic potential to produce enterotoxin genes, as shown in Table 2 and Figure 2. All isolates produced the Sed enterotoxin gene followed by Sea and See at percentages of 70% and 50%, respectively. On the other hand, seven strains had Sea and Sed enterotoxin genes, three isolates harbored Sea, See and Sed enterotoxin genes, and two isolates had gene corresponding to Sed and See enterotoxin genes. Seb and Sec genes were not detected in any of the study isolates.
Table 2: Multiplex-PCR and SET-RPLA results for detection of some S. aureus enterotoxins in raw cow milk.
| IsolatesNo. | Enterotoxin genotyping pattern | Enterotoxin proteina (SET-RPLA) | Antibiotic resistant patternb |
| 1 | Sea, Sed, See | SEA, SED | OXA-AM-DA-AX |
| 2 | Sea, Sed, See | SEA, SED | OXA-AM-TE-AX |
| 3 | Sea, Sed, See | SEA, SED | OXA- AM-CN-AX |
| 4 | Sed | SEA, SED | OXA-AM-AX |
| 5 | Sea, Sed | SED | OXA-AM-DA-TE |
| 6 | Sed, See | SEA, SED | OXA-AM-DA-CN-AX |
| 7 | Sea, Sed | SED | OXA-AM-DA-AX |
| 8 | Sed, See | SEE, SED | OXA-AM-DA-TE-AX |
| 9 | Sea, Sed | SEA, SED | OXA-AX-DA-CN |
| 10 | Sea, Sed | SED | OXA-AX-DA-CN |
aall S.aureus enterotoxin proteins significantly associated with S.aureus enterotoxin genes except SEE protein that was non significantly associated with See gene (P=0.068). bDA: clindamycin, TE: tetracycline, CIP: ciprofloxacin,OX: oxacillin ,AMC: amoxicillin/clavulinic acid, Ax: amoxicillin, AM: ampicillin , CN: gentamycin.
S. aureus enterotoxigenic protein
Enterotoxins, extracted from 10 enterotoxigenic S. aureus broth culture, revealed varying results. All isolates produced SED, six strains produced SEA, while none of the strains produced SEB and SEC. Six isolates produced SEA in combination with SED (Table 2). Interestingly, except for SEE, which was not found associated with See gene (P=0.068), we found that all S. aureus enterotoxin proteins were significantly associated with S. aureus enterotoxin genes (P= 0.014).This can be evidenced in Table 2, where isolates No.4 and No.6 produced SEA enterotoxin, while the encoding gene was not detected by multiplex PCR. On the other hand, enterotoxin proteins (SEE) was not produced, whereas, it’s See gene was detected by multiplex PCR (Figure 3).
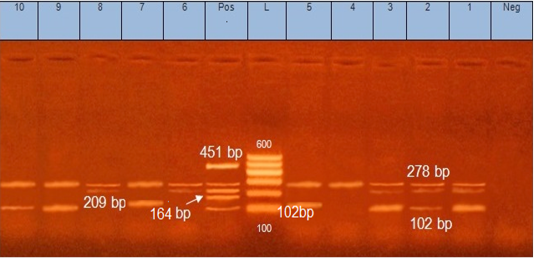
Figure 2: Multiplex PCR for S. aureus enterotoxin genes. Lane (L): 100 bp ladder DNA marker, Lane (Pos): control positive containing DNA of 4 S. aureus strains harboured Seb (164 bp), Sea (102 bp), See (209 bp), Sed (278 bp) and Sec (451 bp) genes. Lane (Neg): control negative. Lanes (1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): isolates positive for Sed gene (278bp). Lanes (1, 2, 3, 5, 7, 9 and 10): isolates positive for Sea gene (102bp). Lanes (1,2, 3, 6and 8): isolates positive for See gene (209bp).
Discussion
Clinically, S. aureus is one of the most significant bacteria that cause food poising worldwide. In this study,20% of raw milk samples from Sharkia Governorate were found contaminated with S. aureus. This rate of prevalence has previously been documented by a number of other studies (Ekici et al., 2004; Santana et al., 2010; Tambekar and Bhutda, 2010; Ammar et al., 2016). They found analyzed samples contaminated with S. aureus in a varying rate of percentages that ranged from 17.34% to 18.80%. Some other studies reported significantly higher levels of contamination ranging from 40% to 61.7 % (Guven et al., 2010; Lingathurai and Vellathurai, 2010; Zakary et al., 2011). Similarly, other studies reported low contamination from 6.2% to 7.3% (Fagundes et al., 2010; Thaker et al., 2013). This is not surprising because milk can be contaminated internally through the production of milk from a diseased animal or externally by an infected person or the surrounding environment (Serraino et al., 2004).
Antimicrobial susceptibility testing of S. aureus against nine different antibiotics showed higher sensitivity rates to ciprofloxacin and vancomycin. These observations are in accordance with those stated previously by others (Aydin et al., 2011). The highest resistance rates were observed for ampicillin, amoxicillin, clindamycin, and gentamycin.With a negligible difference, these observations correlate with those reported previously by others (De Oliveira et al., 2000; Guérin-Faublée et al., 2003; Ammar et al., 2016).
All S. aureus isolates were completely resistant to oxacillin (100%), which contradicts previous findings (Thaker et al., 2013), who found only 20% of S. aureus isolates resistant to oxacillin. High percent isolation of MRSA in this study was considered an extension of previous studies conducted by other researcher groups in Egypt (Ammar et al., 2016) and Bangladesh (Nusrat et al., 2015).
Our findings showed that all the isolates were resistant to at least three antibiotics which is not consistence to previously reported result (Ombui et al., 2000) and contradict several studies which found high levels of MDR in MRSA isolates (Kérouanton et al., 2007; Ge et al., 2017; Sahibzada et al., 2017; Abdi et al., 2018; Suleiman et al., 2018).
S. aureus has the ability to produce enterotoxins that make the potential risk to public health. Our results demonstrated the detection of Sed (100%) gene among all tested S. aureus isolates from raw milk followed by Sea (70%) and See (50%). Sec or Seb genes were not detected in examined isolates. Similar findings were documented previously by others (Carfora et al., 2015).
In another study, a frequent occurrence of See gene was detected in all isolates of SE (100%) followed by a similar presence of Seb and Sec (33.3%), while none of the isolatehadSea or Sed genes (Mansour et al., 2017). Another study revealed Sea gene the most frequent one (61.8%) followed by See (33.1%), Sed (17.5%), Sec (15.9%), and Seb (13.9%) in the S. aureus (Mathenge et al., 2015). We found a significant association between Sea and Sed enterotoxin genes, which is in agreement with findings reported previously (Serraino et al., 2004; Normanno et al., 2005; Morandi et al., 2007).
Multiplex PCR assay can identify the presence of enterotoxin genes but didn’t evaluate their expression. Therefore, in this study, we compared the multiplex PCR results and RPLA (SEA- SED) for toxin type. The RPLA is usually used as a qualitative assessment for S. aureus enterotoxin with a sensitivity of 0.25–0.5 ng/mL concentration of toxins in examined solutions (Rose et al., 1989; Cretenet et al., 2011).
We found that the frequency of SEA and SED enterotoxins were inconsistent with their corresponding genes. Previous studies indicated SEA as the top contributor (80%) to staphylococcal food poisoning outbreaks, followed by SEE, SED, SEB, and SEC. On the contrary, some researchers found a higher expression of SEC and SEB than SED and SEA (Mathenge et al., 2015).
Varying results among PCR and RPLA are not surprising because similar observation for both assays has previously been documented. For instance, a <5% difference in the results of the RPLA and the PCR assays were observed already by others (Jørgensen et al., 2005; Pinto et al., 2005). Ranging from 15 to 32%, marked discrepancies in results of multiplex PCR and RPLA assays, especially for SEA, have been reported (Zouharova and Rysanek, 2008). Such a discrepancy could be explained by the production of enterotoxin in a quantity that was below the detection limit of the RPLA test.
The RPLA results detected some enterotoxin proteins; however, their enterotoxin genes were not detected by multiplex PCR for the same isolate. This finding is in agreement with the observations illustrated previously (Mansour et al., 2017).
Two strains produced SEA enterotoxin, while its encoding gene was not detected using multiplex PCR. Our findings were justified by others (Fraser and Proft, 2008; Argudín et al., 2010) who reported that, though genes more commonly found in groups, enterotoxin genes may found alone, in the large mobile segments of DNA called mobile genetic elements (MGEs) such as transposons, S. aureus pathogenicity islands (SaPIs), plasmids, prophases and the enterotoxin gene clusters (egc).
Conclusion
This study reported that SED and SEA are common enterotoxins in S. aureus isolates originating from raw milk samples. Therefore, the consumption of raw milk should be considered a potential public health hazard. Rapid and efficient detection of enterotoxigenic S. aureus is necessary for consumer safety, and RPLA is recommended for the detection of S. aureus enterotoxin. As presence of enterotoxin genes in S. aureus isolates not mean production of enterotoxin proteins.
Acknowledgments
We thank Animal Health Research Institute, Egypt, for providing the technical support for this study.
Authors Contribution
All authors contributed equally to this work.
Conflict of interests
The authors have declared no conflicts of interest.
Reference