Advances in Animal and Veterinary Sciences
Research Article
Molecular Epidemiology and Sequencing of Avian Pathogenic Escherichia coli APEC in Egypt
Dalia M. Sedeek*, Mohamed M. Rady, Hanaa S. Fedawy, Nagwa S. Rabie
Poultry Diseases Department, Veterinary Research Division, National Research Centre, Dokki, Giza, Egypt.
Abstract | To identify the virulence genes of avian pathogenic Escherichia coli (APEC) in chickens. Four strains were isolated from the lung, spleen, and liver of eighty freshly dead suspected birds obtained from poultry farms. Therefore, identified biochemically and serologically. Two isolates of O78, one isolate as O1 and one isolate as O26) were identified. Six virulence genes (ompT, hlyF, iroN, iss, iutA, tsh) were isolated. The iroN (50%) virulence gene was detected in two isolates of serogroups O78a and O78b at 553 base pair (bp), while hlyF (100%) virulence gene was detected in all four isolates of E. coli at 450 bp and the ompT (75%) virulence gene was detected in three isolates serogroups O78a, O78b and O1 at 496 bp. The virulence gene iss (25%) was only detected in one isolate serogroup O78b at 266 bp, while virulence gene iutA (100%) was detected in all four isolates at 300 bp. The virulence gene tsh (25%) was only detected in one isolate serogroup O78b at 620 bp. All six virulence factors were detected in Egyptian E. coli isolate serogroup O78b, then made a sequence of three virulence genes (iss, tsh, and iutA) of an APEC serogroup O78b isolate and submitted in gene bank with preliminary accession number MN328734 to MN328736. These sequences were compared to reference strains. A supreme nucleotide identity (>98%) was noticed in comparing the gene sequences of an Egyptian E. coli isolate O78b and genes of reference strains. The three virulence genes of Egyptian isolate E. coli O78b were highly related to genes of reference strains of other countries.
Keywords | E. coli, Isolation, PCR, Virulence genes, Sequencing, Chickens
Received | November 17, 2019; Accepted | March 29,2020; Published | May 01, 2020
*Correspondence | Dalia M. Sedeek, Poultry Diseases Department, Veterinary Research Division, National Research Centre, Dokki, Giza, Egypt; Email: [email protected]
Citation | Sedeek DM, Rady MM, Fedawy HS, Rabie NS (2020). molecular epidemiology and sequencing of avian pathogenic Escherichia coli APEC in Egypt. Adv. Anim. Vet. Sci. 8(5): 499-505.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.5.499.505
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Sedeek et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Avian colibacillosis is one of the most famous diseases causing huge economic losses to the poultry industry overall world as it leads to supreme morbidity and mortality of influenced birds (Schouler et al., 2012; Paixao et al., 2016). It is a localized or systemic infection, described by different syndromes involving septicemia, respiratory disease, yolk sac infection, swollen head syndrome, omphalitis, and cellulitis, or an integration of these syndromes (Omer et al., 2010; Ahmed et al., 2013; De-Carli et al., 2015).
APEC infections are related to stress induced by infections with New Castle disease (ND) virus, infectious bronchitis (IB) virus, infectious bursal disease (IBD) virus, mycoplasma, and environmental influences such as humidity, temperature, dust and ammonia in farms. Moreover, E. coli can pierce through the egg shell when it is contaminated with feces then propagate to chickens during hatching, causing high early chick mortality, as well as the yolk sac infection (Dho-Moulin and Fairbrother, 1999; Kabir, 2010).
APEC is Gram-negative bacterium which of Enterobacteriaceae family and considered to be a member of the extra intestinal pathogenic Escherichia coli (ExPEC) group (De-oliveira et al., 2015; Wang et al., 2015; De-Carli et al., 2015). Several virulence genes were clarified in APEC-encoding adhesions, toxins, resistance to the host serum, iron uptake systems, and auto transporter (Ewers et al., 2004; Li et al., 2005; Schouler et al., 2012).
However, numerous survies have reported that these virulence factors are seldom all present in the same isolate and they could be found either individually or polygenically with varying frequencies in clinical isolates (Delicato et al., 2003; Vandekerchove et al., 2005; Knobl et al., 2008).
Past research had already determined some bacterial genes that code to virulence genes in APEC strains. First, it was identified that 9 (ompT, sitA, iroN, iss, iutA, tsh, fyuA, irp-2, and cvaC) of a sum of 38 studied genes occur in high frequencies in APEC (Rodriguez-Siek et al., 2005). The investigators detected genetic criteria for the classification of E. coli strains: isolates with a minimum of 5 virulence associated genes could be identified as pathogenic (APEC), while those with less than 5 as non-pathogenic E. coli (De-Carli et al., 2015).
This study was applied to detect the APEC-associated virulence genes of E.coli isolates with different serogroups which are responsible for colibacillosis in chickens.
MATERIALS AND METHODS
Sampling identification and isolation of bacteria
In this study, four isolates were isolated from the liver, lung, and spleen of eighty freshly dead chickens suspected in colibacillosis from poultry farms in various regions of Cairo.
Birds that underwent this study were treated according to the institutional animal care and use committee of the National Research Centre.
The liver, lung, and spleen were first cultured on nutrient broth (Oxoid ®, Wade Road
Basingstoke, Hampshire, RG248PW, United Kingdom) aerobically at 37 °C for 18 to 24 h before being sub-cultured on MacConkey agar (Oxoid ®) and Eosin methylene blue agar (EMB) (Oxoid ®) at the conditions described above. For each sample, a single suspected colony was picked up and sub-cultured on nutrient agar overnight at 37 °C. Then, isolates with typical characteristics colonies were identified biochemically using Gram staining, and API20E® system then identified serologically. E. coli isolates were stored in storage agar at 4 °C until use.
DNA extraction
Genomic DNA extraction was accomplished as described by Blanco et al. (2004). Bacterial isolates were, first, sub-cultured overnight at 37 °C in Trypticase Soya Broth before being suspended in 200μL of sterile water. Bacteria were boiled for 10 min to release the DNA, followed by centrifugation at 10,000 rpm/5 min. The supernatant containing DNA was stored at −20 °C until used for polymerase chain reaction (PCR) experiments.
Detection of virulence genes (ompT, hlyF, iroN, iss, tsh, and iutA)
Isolated E. coli strains were investigated for the presence of six virulence genes (ompT, hlyF, iroN, iss, tsh, and iutA) which are associated with colibacillosis. For the detection of virulence genes, genomic DNA was extracted from pure cultures of E. coli grown overnight on the MacConkey agar at 37 °C by using the DNeasy Blood and Tissue Kit (Vivantis ®, Vivantis Technologies Sdn. Bhd. Revongen Corporation Center No.12A, Jalan TP 5, Taman Perindustrian UEP, 47600 Subang Jaya, Selangor Darul Ehsan, Malaysia).
The quality of genomic DNA was checked by gel electrophoresis and measuring absorbance at A260/A280 and A260/A230 ratios using the Maestro Nano spectrophotometer (MaestroGen; Model name: MN-913). The conventional PCR was used to amplify the virulence genes. The primers used for amplification were those described previously Table 1 (Ewers et al., 2005; Radwan et al., 2014) were supplied by Metabion (Germany). The PCR was performed in 25μL volume containing 12.5μL Hot start Taq 2X master mix (Vivantis ®), 1μL each primer, 2μL DNA template, and 8.5μL nuclease-free water. The PCR amplifications were conducted in an applied biosystem 2720 thermal cycler and the cycling conditions were identical for all the samples as follows: 94 °C for 4 min; 35 cycles of 30s at 94 °C, 1 min at 60 °C, and 2 min at 68 °C; and 7 min at72 °C for ompT, hlyF, and iroN genes according to (Ewers et al., 2005; Radwan et al., 2014). While 94 °C for 5 min; 35 cycles of 30 s at 94 °C, 54°C, 72°C and 7 min at 72 °C for iss gene according to (Yaguchi et al., 2007), 94 °C for5 min; 35 cycles of 30 s at 94 °C, 54 °C, 72 °C and10 min at 72 °C for tsh gene according to (Delicato et al., 2003), 94 °C for 5 min; 35 cycles of 30 s at 94 °C, 63 °C, 72 °C and 7 min at 72 °C for iutA gene according to (Yaguchi et al., 2007). The products of PCR were separated by electrophoresis on 1.5% agarose gel (Applichem®, Germany, GmbH) in 1x Tris borate EDTA buffer at room temperature using gradients of 5V/cm. For gel analysis, 15 µl of the products were loaded in each gel slot. Gelpilot 100 bp ladder (Qiagen ®, Germany, GmbH) and a gene ruler 100 bp ladder (Fermentas ®, Germany) were used to determine the fragment sizes. The gel was photographed by a gel documentation system (Alpha Innotech, Biometra) and the data was analyzed through computer software.
Sequencing
Sequencing reactions were performed with purified amplified products of the 6 virulence genes. The PCR product was analyzed using the ABI 3130xl Genetic Analyzer (Applied Biosystems). The sequences of nucleotides in each sample were pooled in a single strand of DNA (consensus)
Table 1: Primers sets for demonstrating of selected virulence genes from (APEC) isolates.
| Genes |
Primer Sequence (5′–3′) |
Amplicon size (bp) | Reference |
| ompT |
F:TCATCCCGGAAGCCTCCCTCACTACTAT R: AGCGTTTGCTGCACTGGCTTCTGATAC |
496 | |
| hlyF |
F: GGCCACAGTCGTTTAGGGTGCTTACC R: GCGGTTTAGGCATTCCGATACTCAG |
450 | |
| iroN |
F: AATCCGGCAAAGAGACGAACCGCCT R:GTTCGGGCAACCCCTGCTTTGACTTT |
553 | |
| iss |
F:ATGTTATTTTCTGCCGCTCTG R:CTATTGTGAGCAATATACCC |
266 | |
| tsh |
F:GGT GGT GCA CTG GAG TGG R:AGT CCA GCG TGA TAG TGG |
620 | |
| iutA |
F:GGCTGGACATGGGAACTGG R:GTCGGGAACGGGTAGAATCG |
300 |
F: forward; R: reverse; bp: base pair.
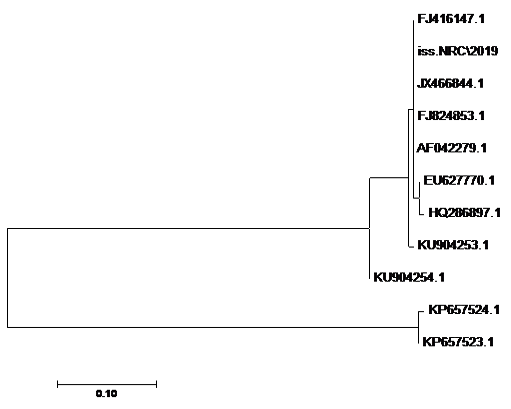
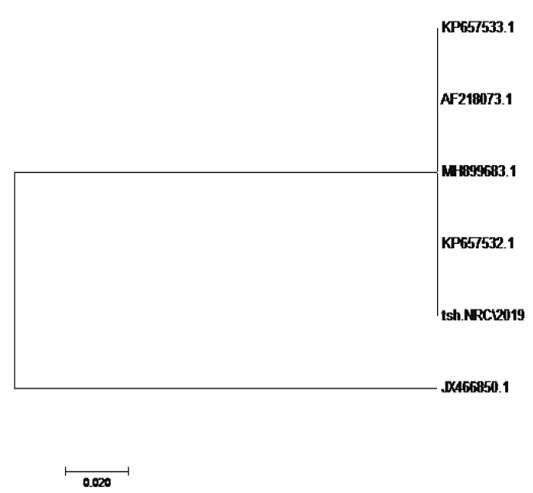
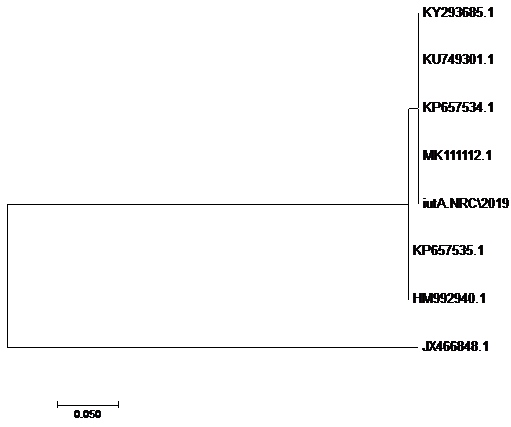
using the SeqMan program. Consensus sequences of each gene were aligned with reference sequences for comparative genetic similarity analysis using MegAlign Program (Lasergene, DNAStar, and Madison, WI). The reference sequences for iss gene were iss genes from the reference Brazilian E. coli strain with accession number FJ824853.1 and KP657523.1 and KP657524.1, reference gene of Iranian E. coli strain with accession number FJ416147.1 and HQ286897.1, reference gene of American E. coli strain with accession number AF042279.1, reference gene of Chinese E. coli strains with accession number JX466844.1 and EU627770.1 and reference gene of Egyptian E. coli strains with accession number KU904253.1 and KU904254.1. While the reference sequences for tsh gene were tsh genes from the reference Brazilian E.coli strain with accession number KP657532.1 and KP657533.1, reference gene of American E. coli strain with accession number AF218073.1, reference gene of Canadian E. coli strain with accession number MH899683.1 and reference gene of Chinese E.coli strain with accession number JX466850.1 and the reference sequences for iutA gene were iutA gene from the reference Brazilian E.coli strain with accession number KP657534.1 and KP657535.1, reference gene of Russian E.coli strain with accession number KY293685.1,reference gene of the Iranian E.coli strain with accession number KU749301 and MK111112.1, reference gene of Indian E.coli strain with accession number HM992940.1 and reference gene of the Chinese E. coli strains with accession number JX466848.1.
The nucleotide sequence data reported in this paper were submitted to the Gene Bank nucleotide sequence database and have been assigned with the preliminary accession numbers MN328734, MN328735, and MN328736 with designation iss. NRC/2019, tsh.NRC/2019 and iutA.NRC/2019, respectively.
Results and Discussion
Detection and sequences of virulence genes
In the present study, APEC was recovered and diagnosed from suspected cases of diseased chickens. Suspected samples were cultivated under aseptic conditions. Lactose fermenting colonies were detected on the MacConkey agar and characteristic green metallic sheen on EMB agar. Biochemically and serologically identification revealed that four isolates were identified as E. coli, two isolates of serogroup O78, one isolate of serogroup O1 and one isolate of serogroup O26.
All isolates were investigated by molecular technique to detect the presence of six virulence-associated genes described for APEC as ompT (episomal outer membrane protease), hlyF (putative avian hemolysin), IroN (salmochelin siderophore receptor), iss (the episomal increased serum survival), tsh (temperature-sensitive hemagglutinin gene) and iutA (ferric aerobactin receptor gene) associated to field isolates of E. coli.
Our result demonstrated that the virulence gene hlyF (100%) was detected from all the four APEC isolates (O78a, O1, O26, and O78b) at amplicon size 450 bp. The virulence gene iroN (50%) was detected from two APEC isolates (O78a and O78b) at amplicon size 553 bp. The virulence gene ompT (75%) was detected from three APEC strains (O78a, O1, and O78b) at amplicon size 496 bp as shown in Figure 1. The virulence gene tsh (25%) was only detected from one isolate (O78b) at amplicon size 620 bp as shown in Figure 2. The virulence gene iutA (100%) was detected from all four isolates (O78a, O1, O26 and O78b) at 300 bp, while the virulence gene iss (25%) was only detected from one isolate (O78b) at amplicon size 266 bp. Figure 3.
From these results we can conclude that the serogroup O78a has four virulence genes (iroN, hlyF, ompT, and iutA) whenever, the serogroup O78b has six virulence genes (iroN, hlyF, ompT, iss, tsh, and iutA) while, the serogroup O1 has three virulence genes (hlyF, ompT, and iutA) and serogroup O26 has two virulence genes (hlyF and iutA) as shown in Table 2.
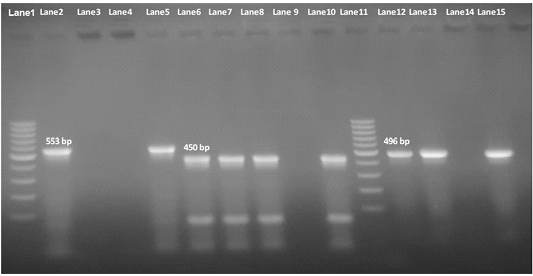
Figure 1: Agarose Gel Electrophoresis (AGE) showing PCR Products of E.Coli different genes. Lane 1 and Lane 11: 100 bp marker; Lane 2, 3, 4 and 5: 553 bp PCR Products of E. coli iroN gene; Lane 6,7, 8 and 10: 450 bp PCR Products of E. coli hlyF gene; Lane 12, 13, 14and 15: 496 bp PCR Products of E. coli ompT gene; NB: iroN gene not included in both O1 and O26 E .coli isolates (Lane 3 and 4 are negative), as well as ompT not included in O26 E. coli in O2 E. coli isolate e(Lane 14 is negative).
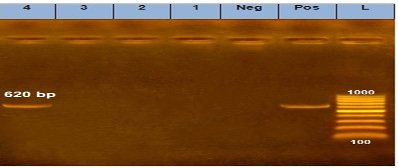
Figure 2: Agarose Gel Electrophoresis (AGE) showing PCR Products of E. coli different genes: Lane (L): 100 bp marker. Lane 1, 2, 3 and 4: 620 bp PCR products of E. coli tsh gene.
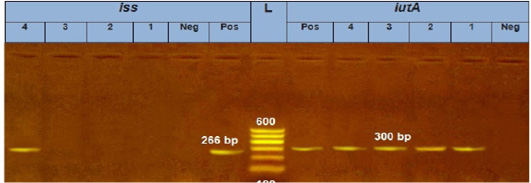
Figure 3: Agarose Gel Electrophoresis (AGE) showing PCR Products of E. coli different genes: Lane (L): 100 bp marker; Lane 1, 2, 3 and 4 (left): 266 bp PCR products of E.coli iss gene; Lane 1, 2, 3 and 4 (Right): 300 bp PCR products of E.coli iutA gene.
The sequences of amplified products of (iss, tsh, and iutA) genes that obtained from O78b isolate were detected then compared with the reference genes in the gene bank. The results showed a high degree of identity (>98%) to every gene with the sequences of respective coding amino acid. Several nucleotide sequences were synonymous and the respective proteins of amino acid sequences were matched. There were non-synonymous mutations in iutA gene of O78b isolate, with only two amino acid modifications in the sequence of one gene (iutA). Phylogenetically iss gene (iss.NRC/2019) with accession no. MN328734 of an Egyptian isolate E. coli O78b related to the cluster of iss gene of E. coli strains with accession no. JX466844.1, FJ824853.1, AF042279.1, HQ286897.1, and EU627770.1 not related to cluster with accession no. KP657523.1, KP657524.1, and KU904254.1. Figure 4 While tsh gene (tsh.NRC/2019) with accession no. MN328735 of an Egyptian isolate E. coli O78b related to the cluster of tsh gene of E. coli strains with accession no. MH899683.1, AF218073.1, KP657532.1, and KP657533.1 not related to cluster with accession no. JX466850.1. Figure 5 While iutA gene (iutA.NRC/2019) with accession no. MN328736 of an Egyptian isolate E. coli O78b related to the cluster of iutA gene of E. coli strains with accession no. MK111112.1, KY293685.1, KU749301 and KP657534.1 not related to cluster with accession no HM992940.1, KP657535.1 and JX466848.1. Figure 6.
Table 2: Result of virulence genes associated to field isolates of E. coli.
| Isolate no. |
Isolate serogroup |
Results of genes containing in each isolate | |||||
| iroN | hlyF | ompT | iss | iutA | tsh | ||
| 1 |
O78a |
+ | + | + | - | + | - |
| 2 | O1 | - | + | + | - | + | - |
| 3 | O26 | - | + | - | - | + | - |
| 4 |
O78b |
+ | + | + | + | + | + |
E. coli serogroup O78b isolate.
In countries around the globe, poultry is of considerable importance as a vital route out of poverty, ensuring food and sustainable livelihoods. The poultry industry has changed significantly and continues to develop and enlarge all over the world as well as in Egypt. Avian colibacillosis is considered one of the most famous diseases causing great losses economically in the poultry industry overall the world (Schouler et al., 2012; Paixao et al., 2016). APEC has the virulence genes responsible for colibacillosis disease (Barnes et al., 2008). These virulence genes posses the essential molecular markers for APEC strains characterization (Johnson et al., 2008).
The ability of APEC for pathogenicity is associated with broad range of virulence agents which are coded by virulence-associated genes. Detection of at least five virulence genes represent the pathogenicity of APEC strain and the most important virulence genes in APEC (ompT, iutA, iss, tsh, papC, iucD, hlyF, irp-2, iron, cva/cvi, and astA). In our study, four field isolates of E. coli were recovered and diagnosed according to biochemical and serological identification as two isolates of serogroup O78, one isolate of serogroup O1 and one isolate of serogroup O26. Numerous virulence genes were declared in APEC encoding toxins, adhesins, iron uptake systems, resistance to the host serum, and auto transporter (Ewers et al., 2004; Schouler et al., 2012). We assessed the presence of six well-characterized virulence genes in four E. coli isolates, the salmochelin siderophore receptor (iroN) is invasive related genes which is responsible for uptaking (Fe) ions obtained from the body host. The episomal outer membrane protease (ompT) and episomal increased serum survival (iss) both are related to protectins/serum resistance genes which are responsible for resistance to innate immunity and increased survival of bacteria in host serum, respectively. The putative avian hemolysin (hlyF) is toxin related genes and responsible for making pores in host cells membranes. The ferric aerobactin receptor precursor (iutA) is one of the genes related to iron uptaking system which has sidrophore uptake transmembrane transporter activity. Temperature sensitive hemagglutinin (tsh) is adhesion related genes which has both agglutinin and protease activity (Johnson et al., 2008; Ahmed et al., 2013; de Oliveira et al., 2015; Sarowska et al., 2019).
Our result demonstrated that the virulence gene iroN was detected from two isolates (O78aand O78b) (50%) at mplicon size 553 bp.The virulence gene hlyF was detected from all the four isolates (100%) at amplicon size 450 bp. The virulence gene ompT was detected from three isolates (2 isolate O78a, O78b, and O26) (75%) at amplicon size 496 bp as shown in Figure 1.
The virulence gene tsh was only detected from one isolate (25%) (serogroup O78b) at amplicon size 620 bp as shown in Figure 2.
The virulence gene iutA was detected from all four isolates (100%) at 300 bp, while the virulence gene iss was only detected from one isolate (O78b) (25%) at amplicon size 266 bp.as shown in Figure 3.
We can conclude that the strain (O78b) has six virulence genes (iroN, hlyF, ompT, iss, tsh, and iutA) while, the strain (O78a) has four virulence genes (iroN, hlyF, ompT, and iutA). On the other hand, strain (O1) has three virulence genes (hlyF, ompT, and iutA). Strain (O26) has two virulence genes (hlyF and iutA) as shown in Table 2.
The pathogenicity of APEC strain is evaluated by the recording of at least five virulence genes according to the genetic criteria, which enable them to survive an extraintestinal life (De Carli et al., 2015). The virulence genes iss, papC, and tsh were not found in the non-APEC isolates. The author had reported high incidence of virulence associated genes (hlyF, 100%; ompT, 100%; iroN, 98. 8%; iss, 96.3%; and iutA, 81.5%) within the APEC strains (De Carli et al., 2015).
In another study, iutA, astA, and iucD virulence genes, are established as basic genes responsible for E. coli pathogenicity. This study also showed that there was no uniform and absolute combination of the virulence genes which can distinguish APEC and non-APEC strains of E. coli. Moreover, the detection of iss, tsh and papC genes exclusively only in the APEC strains could be reliabled as colibacillosis important virulent agents (Mohamed et al., 2018; Subedi et al., 2018).
Additionally, nucleotide and amino acid sequences analyzed in this survey were highly similar (>98%) to both corresponding genes of the reference strains as shown in Figure 4, 5 and 6 (Skyberg et al., 2006; Johnson et al., 2007). Although these genes are conserved and no polymorphic, they are rarely all found among an isolate. APEC strains are considered as a heterogeneous bacteria. There is a specific combination of genes for each APEC strain to cause colibacillosis in poultry (Schouler et al., 2012). There are essential virulence genes which are related to the presence of a plasmid similar to pColV in different studies recoreded overall the world (Jeong et al., 2012; Schouler et al., 2012; Dissanayake et al., 2014).
Acknowledgment
The authors thank alot the National Research Centre for funding this article (Grant number: AR111213).
Authors Contribution
DALIA M. SEDEEK designed the study, drafted and revised the manuscript, MOHAMED M. RADY, HANAA S. FEDAWY and NAGWA S. RABIE shared in samples collection, performing the tests, manuscript writing and data analysis. All authors read and approved the final manuscript.
Statement of conflict of interest
There is no conflict of interest.
References