Advances in Animal and Veterinary Sciences
Research Article
Virulence Gene Constellations Associated with Lethality in Avian Pathogenic E. coli Recovered from Broiler Chickens
Ahmed Ali1*¥, Ahmed I. Abd El-Mawgoud2¥, Al-Hussien M. Dahshan1, Azza A. EL-Sawah1, Soad A. Nasef3, Mahmoud Ibrahim4
1Poultry Diseases Department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef 62511 Egypt; 2Reference Laboratory for Veterinary Quality Control on Poultry Production, Animal Health Research Institute, Fayoum Branch, Egypt; 3Reference Laboratory for Veterinary Quality control on Poultry Production, Animal Health Research Institute, Dokki, Giza, Egypt; 4Department of Birds and Rabbit Medicine, Faculty of Veterinary Medicine, University of Sadat City, Menoufiya, Egypt.
Abstract | In this study, avian pathogenic E. coli (APEC) isolated from broiler chickens in Fayoum and Beni-Suef governorates in Egypt were morphologically and biochemically identified. Selected isolates were serologically identified, and their antimicrobial susceptibility profiles were determined. Ten isolates from the most predominant serogroups were subjected to virulence and antimicrobial resistance genes detection using PCR. In vivo pathogenicity test was performed for 5 E. coli strains that were selected based on serogroups prevalence, virulence, and antimicrobial genes patterns. Results revealed an incidence of 53.8% of E. coli from broiler chickens with the predominance of O125 (30%), O119 (20%), O126, and O86a (15% each) serogroups. The selected 20 E. coli strains were multi-drug resistant (MDR) but sensitive to fosfomycin. The β-lactams and tetracyclines resistance genes were detected in all tested isolates, however, aminoglycoside and quinolone resistance genes were not detected. In this study, 10% of isolates were resistant to colistin and the mcr-1 gene was detected by PCR. In vivo pathogenic strains consistently harbored the virulence gene pattern of fimH, fimA, papC, iutA, and tsh which was the most common gene constellation detected. The detection of tsh gene was consistently associated with lethality in day-old chicks. In conclusion, the current study demonstrated the high prevalence of multidrug-resistant E. coli in broiler chickens representing a potential public health concern. Additionally, the virulence gene constellation of fimH, fimA, papC, iutA, and tsh was found to be associated with lethality in day-old chicks and can be used as a virulence marker of APEC.
Keywords | E. coli, Broiler chickens, MDR, Virulence, Pathogenicity, Egypt
Received | May 25, 2019; Accepted | Novemebr 10, 2019; Published | November 25, 2019
*Correspondence | Ahmed Ali, Poultry diseases department, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef 65211, Egypt; Email: [email protected]
Citation | Ali A, El-Mawgoud AIA, Dahshan AM, EL-Sawah AA, Nasef SA, Ibrahim M (2019). Virulence gene constellations associated with lethality in avian pathogenic E. coli recovered from broiler chickens. Adv. Anim. Vet. Sci. 7(12): 1076-1082.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.12.1076.1082
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Ali et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Avian pathogenic Escherichia coli (E. coli) (APEC), belonging to the extra-intestinal pathogenic Escherichia coli (ExPEC) causes many forms of poultry colibacillosis including, localized and systemic infections including respiratory origin colisepticemia, enteric origin colisepticemia and localized infections such as; cellulitis, omphalitis, diarrhea and swollen head syndrome (Ewers et al., 2009). Though E. coli colonizes the lower digestive tract of chicks within 24hr after hatching (Ballou et al., 2016), their pathogenicity is generally enhanced or initiated by predis posing factors, such as environmental factors, viral infections, mycoplasma infections, and immune-suppression (Bopp et al., 2003). In Egypt; several studies showed a high prevalence of E. coli among poultry farms especially in broiler chickens from living diseased and freshly dead chickens (Abd-ElTawab et al., 2015; Ali et al., 2019).
Based on the detection of five virulence genes (namely hlyF, iutA, iroN, iss, and ompT) a pentaplex panel was proposed as minimal criteria to distinguish between APEC strains and avian fecal E. coli isolates (Johnson et al., 2008). However, another study proposal depending on both the numbers and the combination patterns of virulence-associated genes suggested that highly pathogenic E. coli strains harbored at least 8 to 13 virulence genes while intermediate pathogenic strains harbored at least 5 to 8 virulence genes (Wang et al., 2015).
Phenotypic characterization of APEC strains is also widely used to correlate the virulence criteria and the pathogenicity of isolated E. coli strains. Expanded spectrum beta-lactamases (ESBL) Enterobacteriaceae incidence is increasing among clinical isolates in animals posing a zoonotic risk to humans. Considering the unorganized large-scale use of antimicrobials in Egypt, it was not surprising that researchers detected ESBL in poultry and other animals (El-Shazly et al., 2017). Recently, the plasmid-mediated colistin resistance gene (mcr-1) detection is increasing in Asia (Liu et al., 2016) and Europe (Hasman et al., 2015). The first report of mcr-1 in a clinical human isolate from Egypt and Africa was in 2016 (Elnahriry et al., 2016) following the first reports in animals in Algeria (Olaitan et al., 2016) and Egypt (Khalifa et al., 2016). The high prevalence of mcr-1 in animal isolates compared to human clinical isolates is a strong proof that the antimicrobials misuse in livestock is probably increasing the risk of zoonotic transmission of these bacteria from animals to humans (Zhao et al., 2018).
The aim of this study was to determine the distribution of both virulence and resistance genes in APEC isolates recovered from broiler chickens. Additionally, the relationship of the distribution and combinations of virulence-associated genes to the in vivo characterization of the isolated APEC was studied.
MATERIALS AND METHODS
A total of 400 samples of internal organs were collected from moribund broiler chickens suffering from colibacillosis with pericarditis, perihepatitis, and air-saculitis from 20 farms in Fayoum and Beni-Suef governorates during the period between December 2017 to November 2018. For each broiler farm, 20 organ samples (heart, liver, lung and air sacs, spleen) were collected, labeled and transported in icebox to the Reference Laboratory of Quality Control on Poultry Production, Animal Health Research Institute (RLQP, Giza, Egypt) for analysis.
Bacteriological isolation and identification
The samples were processed under aseptic techniques. Briefly, one gram of the sampled organ was inoculated with 9 ml of buffered peptone water and incubated at 37°C overnight. To identify E. coli, a loopful of broth was streaked on MacConkey agar. Isolated bacteria were then morphologically and biochemically identified (Quinn et al., 2011). Congo red binding assay (Berkhoff and Vinal, 1989) and hemolytic activity on blood agar (Marilda et al., 1990) were conducted to initially differentiate between pathogenic and non-pathogenic bacterial isolates of E. coli and for detection of hemolytic and non-hemolytic isolates. Based on morphological and biochemical and initial pathogenicity identification, 20 isolates from the investigated 20 broiler farms were selected and serologically identified according to (Starr, 1986).
Antimicrobial susceptibility test
The 20 serogrouped E. coli were tested against a panel of 15 antimicrobial agents by using the disc diffusion method and the results were interpreted (CLSI, 2017). The following antimicrobial agents were used; amoxicillin (10µg), ampicillin (10µg) amoxicillin plus clavulanic acid (30µg), florfenicol (30µg), ciprofloxacin (5µg), doxycycline hydrochloride (30µg), oxytetracycline (30µg), cefotaxime sodium (30µg), cefepime (30µg), streptomycin (10µg), gentamicin (10µg), kanamycin (30µg), fosfomycin (200µg) sulpha-trimethoprim (25µg) and colistin sulfate (10µg) (Oxoid Ltd., Hampshire, UK). E. coli ATCC®8379 (Microbiologics®, St Cloud MN, USA) was used as a control organism.
Detection of the virulence and resistance encoding genes in E. coli isolates
For molecular characterization, 10 E. coli isolates representing the most predominant serogroups in addition to an O78 strain were selected. Bacterial DNA was extracted from selected colonies using the QIAamp DNA mini kit (Qiagen) according to the manufacturer’s instructions. Oligonucleotide primers used for virulence and antimicrobial resistance genes detection are shown in supplementary Table 1.
In vivo pathogenicity test
Chicken experiments were conducted according to Animal Research Ethics Guidelines at the faculty of veterinary medicine, Beni-Suef University, Egypt. To evaluate the relationship between the pattern of both virulence and antimicrobial susceptibility of the pathotypic properties of isolated E. coli, the in vivo pathogenicity test was performed. Five E. coli strains were selected based on serotypes prevalence, virulence genes, and antimicrobial genes pattern, and zoonotic importance (namely; S82-O125, H6-O119, H73-O126, H91-O86a, and S54-O78). For each strain, 5 groups of day-old chicks (no. 5 chicks/group) were inoculated subcutaneously with 0.5 ml of bacterial suspension approximately containing 108 CFU/ml. Chicks were monitored for 4 days post-inoculation, clinical signs and mortality were recorded. The strains were classified as pathogenic when at least one chick died (Schouler et al., 2012). A group of five one-day-old SPF chicks was used as saline inoculated negative control group.
Table 1: The isolation rate of E. coli in different organs from Fayoum and Beni-Suef governorates.
| Organ sample (100 each) | Fayoum governorate no. (%) | Beni-Suef Governorate no. (%) | Total isolates/organs % |
| Heart and heart blood | 29(58%) | 30 (60%) | 59% |
| Liver | 29 (58%) | 22 (44%) | 51% |
| Lung /air sac | 27 (54%) | 26 (52%) | 53% |
| Spleen | 24 (48%) | 28 (56%) | 52% |
| Total | 109 (54.5%) | 106 (53.0%) | 53.8% |
Results
Prevalence of E. coli in the study area
Out of 400 samples, 329 gram-negative lactose fermenter bacilli were identified on Macconkey agar. Biochemical identification revealed the identification of 215 E. coli strains with an incidence of 53.8%. No significant differences between the isolation rates of E. coli any of the selected organs with the highest isolation rate from heart and heart blood (59%) (Table 1). All identified strains were able to bind Congo red, however, 16.28% of them have hemolytic activity (data not shown).
The serogrouping of the selected 20 E. coli isolates recovered from diseased chickens revealed a high prevalence of E. coli O125 (30%), O119 (20%), O126, and O86a (15% each). Other serogroups included O78, O158, O20, and O142 (5% each).
Antimicrobial susceptibility testing
In vitro sensitivity tests revealed that most of the selected 20 E. coli are highly sensitive to fosfomycin (85%) followed by colistin sulfate (40%). In contrast, most E. coli isolates were highly resistant to cefepime, florfenicol (95%) followed by amoxicillin, ampicillin, oxytetracycline, and streptomycin (90%) (Supplementary Figure 1). Multi-drug resistance (MDR) was detected amongst the twenty E. coli isolates (100%) indicating the resistance for ≥3 antibiotic classes (Table 2).
Detection of resistance and virulence genes
Antimicrobial resistance genes
The β-lactams resistance gene (blaTEM) and the tetracyclines resistance (tetA) genes were detected in all tested isolates, followed by sulfonamides resistance gene (sul1) which was detected in 9 out of 10 isolates. Three trimethoprim, phenicol, and colistin (mcr-1) resistance genes were detected in 8/10 isolates. The blaCTX-M-1 gene was detected in only 10% of isolates and two genes weren’t detected in any of the examined isolates which were aminoglycoside (aadA1) and quinolones (qnrA) resistant. The number of resistance genes detected per isolate ranged from 2 genes to 7 genes in one E. coli isolate (Table 3, Figure 1).
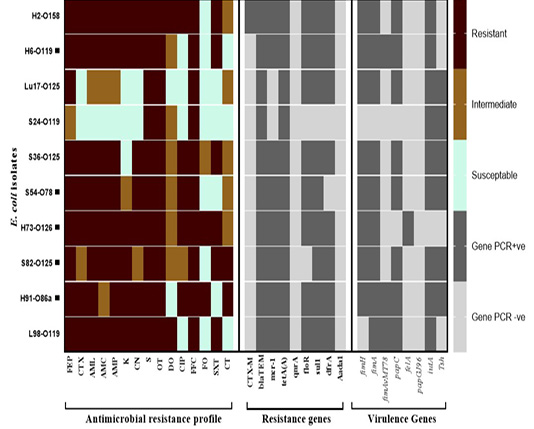
Figure 1: Antimicrobial resistance and virulence genes constellation in the selected E. coli isolates recovered from broiler chickens. Abbreviations; FEP: cefepime, CTX: cefepime, AML: amoxicillin, AMC: amoxicillin/clavulanic acid, AMP: ampicillin, K: Kanamycin, CN: gentamycine, S: streptomycin, OT: oxytetracycline, DO: doxycycline, CIP: ciprofloxacin, FFC: florfenicol, FO: fosfomycin, SXT: sulphamethoxazol-trimethoprim, CT: colistin sulphate. Black square (▄) denotes for the strains tested for their in vivo pathogenicity.
Virulence genes
Out of 8 genes investigated, the number of genes detected per isolate ranged from as few as 2 genes (one isolate, 10%) to as many as 6 genes (one isolate, 10%). The type 1 fimbriae gene (fimA) and iron acquisition gene (iutA) were the most commonly detected genes (9/10 isolates) followed by the fimH gene and papC genes (8/10). The temperature-sensitive hemagglutinin gene (tsh) was detected only in 6/10 isolates. Other genes detected include the fimAvMT78 and felA in 3/10 and 1/10 isolates, respectively. None of the tested isolated harbored the papG gene (Table 3, Figure 1).
In vivo pathogenicity test.
According to the pathogenicity test, 3 strains were found as APEC and caused the death of all chicks or 3 out of 5 inoculated chicks (strains S54-O78and S82-O125, and strain H91-O86a, respectively) (Table 4).
Table 2: Antimicrobial resistance pattern and multi-drug resistance (MDR) indices of E. coli isolated from broiler chickens.
| Strain code |
Antimicrobial resistance pattern A |
Number of antimicrobial classes in pattern |
MDR Index B |
|
H2-O158 |
FEP-CTX-AML-AMC-AMP-K-CN-S-OT-DO-CIP-FFC-SXT | 6 | 0.866 |
|
H6-O119 ▄ |
FEP-CTX-AML-AMC-AMP-K-CN-S-OT-FFC-SXT | 5 | 0.733 |
|
S13-O86a |
FEP-AML-AMC-AMP-S-CIP-FFC-SXT | 5 | 0.533 |
|
Lu17-O125 |
FEP-S-OT-FFC | 4 | 0.266 |
|
S24-O119 |
S-OT-FFC | 3 | 0.200 |
|
Lu30-O119 |
FEP-AML-AMC-AMP-K-CN-S-OT-CIP-FFC-SXT | 6 | 0.733 |
|
L35-O125 |
FEP-CTX-AML-AMC-AMP-CN-S-OT-FFC-SXT | 5 | 0.666 |
|
S36-O125 |
FEP-CTX-AML-AMC-AMP-CN-S-OT-CIP-FFC-SXT | 6 | 0.733 |
|
Lu42-O126 |
FEP-CTX-AML-AMC-AMP-K-S-OT-CIP-FFC | 5 | 0.666 |
|
L48-O126 |
FEP-AML-AMC-AMP-S-OT-FFC-SXT | 5 | 0.533 |
|
S54-O78 ▄ |
FEP-CTX-AML-AMC-AMP-CN-S-OT-CIP-FFC | 5 | 0.666 |
|
Lu58-O142 |
FEP-AML-AMC-AMP-K-CN-OT-CIP-FFC-SXT | 6 | 0.666 |
|
L62-O20 |
FEP-AML-AMC-AMP-S-OT-FFC-SXT | 5 | 0.533 |
|
L70-O125 |
FEP-AML-AMC-AMP-OT-DO-CIP | 3 | 0.466 |
|
H73-O126 ▄ |
FEP-CTX-AML-AMC-AMP-K-CN-S-OT-CIP-FFC-FO-SXT | 7 | 0.866 |
|
L76-O125 |
FEP-CTX-AML-AMC-AMP-CN-S-OT-DO-CIP-FFC | 5 | 0.733 |
|
S82-O125 ▄ |
FEP-AML-AMC-AMP-K-S-OT-FFC-SXT-CT | 6 | 0.666 |
|
Lu90-O86a |
FEP-CTX-AML-AMC-AMP-K-CN-S-CIP-FFC-SXT | 5 | 0.733 |
|
H91-O86a ▄ |
FEP-CTX-AML-AMP-K-CN-S-OT-CIP-FFC-FO-CT | 7 | 0.800 |
|
L98-O119 |
FEP-CTX-AML-AMC-AMP-K-CN-S-OT-DO-FFC-SXT | 5 | 0.800 |
A Resistance to fifteen selected antimicrobials, as determined by disc diffusion assay. FEP: cefepime, CTX: cefotaxime, AML: amoxicillin, AMC: amoxicillin/clavulanic acid, AMP: ampicillin, K: kanamycin, CN: gentamycin, S: streptomycin, OT: oxytetracycline, DO: doxycycline, CIP: ciprofloxacin, FFC: florfenicol, FO: fosfomycin, SXT: sulphamethoxazol/trimethoprim, CT: colistin sulphate; Ban isolate was defined as being multidrug resistant (MDR) if it was resistant to ≥ 3 antimicrobial classes; Black square (▄) denotes for the strains tested for their in vivo pathogenicity.
Table 3: Resistance and virulence genes patterns of E. coli isolated from broiler chickens.
| Resistance gene pattern | no. (%) | Virulence gene pattern | no. (%) |
| 7 genes | 6 genes | ||
|
blaTEM, tetA(A), sul1, floR, dfrA, mcr-1, blaCTX-M-1 |
1(10) | fimH, fimA, fimAvMT78, iutA, tsh, papC | 1(10) |
| 6 genes | 5 genes | ||
|
blaTEM, tetA(A), sul1, floR, dfrA, mcr-1 |
5(50) | fimH, fimA, iutA, tsh, papC | 4(40) |
| 5 genes | fimH, fimA, fimAvMT78, iutA, papC | 1(10) | |
|
blaTEM, tetA(A), sul1, floR, dfrA |
1(10) | 4 genes | |
|
blaTEM, tetA(A), sul1, floR, mcr-1 |
1(10) | fimH, fimA, iutA, papC | 1(10) |
|
blaTEM, tetA(A), sul1, dfrA, mcr-1 |
1(10) | fimA, fimAvMT78, iutA, papC | 1(10) |
| 2 genes | 3 genes | ||
|
blaTEM, tetA(A) |
1(10) | fimH, fimA, felA | 1(10) |
| 2 genes | |||
| iutA, tsh | 1(10) |
DISCUSSION
In the current study, the prevalence of APEC was investigated in broiler chickens from two governorates in Egypt. The prevalence of APEC was 53.8 % with the highest isolation rate from heart and heart blood (59%) that proposed pericarditis as being a characteristic of colisepticemia (Lister, 2008). The preliminary phenotypic
Table 4: In vivo pathogenicity test for the selected E. coli strains.
|
Days post inoculation |
Strain code | ||||||
|
H6-O119 |
S54-O78 |
H73-O126 |
S82-O125 |
H91-O86a |
Negative | ||
| Day 1 | Sick | 2 | 1 | 2 | 1 | 4 | 0 |
| Dead | 0 | 4 | 0 | 4 | 1 | 0 | |
| Day 2 | Sick | 1 | 0 | 1 | 1 | 2 | 0 |
| Dead | 0 | 1 | 0 | 0 | 2 | 0 | |
| Day 3 | Sick | 1 | - | 1 | 0 | 2 | 0 |
| Dead | 0 | - | 0 | 1 | 0 | 0 | |
| Day 4 | Sick | 1 | - | 1 | - | 2 | 0 |
| Dead | 0 | - | 0 | - | 0 | 0 | |
| Day 5 | Sick | 1 | - | 1 | - | 2 | 0 |
| Dead | 0 | - | 0 | - | 0 | 0 | |
| Total Dead no. (%) | 0/5 (0%) | 5/5 (100%) | 0/5 (0%) | 5/5 (100%) | 3/5 (60%) | 0/5 0% | |
|
PathotypeA |
NP | P | NP | P | P | -- | |
A Pathotype defined as; NP: non-pathogenic and P: pathogenic E. coli.
characterization utilizing the Congo red binding test and hemolytic activity revealed that all isolated E. coli (215 isolates) were able to bind Congo red, however, only 16.28% of E. coli isolates showed hemolytic activity. Hence, we selected 20 representative isolates to be further serogrouped. The E. coli of O125, O119, O126, and O86a were the most predominant serogroups. Though variable E. coli serotypes in different geographic areas were detected (Ibrahim et al., 2019), more or less similar results were reported from commercial poultry in Egypt (Ezzeldeen et al., 2010; Ramadan et al., 2016; Ali et al., 2019).
Antibiotic susceptibility testing was conducted for the 20 serogrouped E. coli isolates with 15 antibiotics including those used in human and veterinary medicine. Most of the selected 20 E. coli are highly sensitive to fosfomycin (85%) which is a promising candidate for the treatment of infections in veterinary medicine (Pérez et al., 2014). Colistin is one of the drugs of choice for the treatment of MDR Gram-negative bacteria in humans (Lv et al., 2018), in this study 10% of isolates were resistant to colistin. On the other hand, 8 out of 10 tested E. coli isolated were positive for mcr-1 PCR including 2 phenotypically resistant, 4 intermediately susceptible and 2 susceptible isolates to colistin. The emergence of mcr-1 associated with colistin resistance in E. coli is increasing in Egypt and worldwide (Lima Barbieri et al., 2017). These results further confirm that poultry production practices could result in a potential source of resistant bacteria to the human. A recent study demonstrated the sharing of serotypes, virulence genes, and phylotypes between human and APEC isolates (Ramadan et al., 2016).
Unexpectedly, 95% of E. coli isolates were resistant to cefepime (4th generation cephalosporines) despite it is not approved for use in poultry by the authorities. This may be attributed to the continuous use of other cephalosporines and the presence of cross-resistance and/or the abusive administration of several antibiotics for infection or prophylaxis especially in illegal poultry farms in Egypt. As expected, amoxicillin, ampicillin, oxytetracycline, and streptomycin resistant E. coli represented 80-90% of tested isolates (Younis et al., 2017) with the detection of the β-lactams and other corresponding resistance genes associated with phenotypic resistance profiles. Though the blaCTX-M-1 gene was detected in only 1 E. coli isolate, it is worthy to note that more than 90 different CTX-M β-lactamases genes are documented. The variety and the increased prevalence of blaCTX-M-1 is a serious risk for the clinical usage of third-generation cephalosporins (Randall et al., 2012). In contrast to phenotypic characterization, two genes were not detected in any of the examined isolates; the aminoglycoside and the quinolones resistance genes (aadA and qnrA, respectively). This contradiction is probably due to that the phenotypic resistance to these 2 antimicrobial categories is known to be mediated with other genes of resistance (e.g strA and strB for aminoglycoside and gyrA for quinolones resistance ) (Xie et al., 2014).
Detection of virulence genes markers associated with the lethality of APEC isolates was proposed as a diagnostic tool for identifying avian pathogenic E. coli (Dozois et al., 2000). Hence, we tested the pathogenicity of 5 molecularly characterized predominant serogroups E. coli isolates with different virulence gene constellations. Three out of five isolates were found pathogenic in day-old chicks. The pathogenic strains consistently harbored the virulence gene pattern of fimH, fimA, papC, iutA, and tsh which was the most common gene constellation detected. Additionally, the temperature-sensitive hemagglutinin (tsh) gene was the most important virulence marker of APEC as its absence remarkably diminish the pathogenicity of the E. coli isolates. It is worthy to note that the tsh gene exhibits different virulence functions and contain proteases, adhesins, cytotoxins, and cell invasion proteins (Henderson et al., 1998). The strong association with internal organs colonization, septicemia, and lethality in day-old chicks (Ngeleka et al., 2002), probably make it a useful target for pathotyping of APEC.
In conclusion, the current study demonstrated the high prevalence of multidrug-resistant E. coli in broiler chickens in Egypt. An increased detection rate of the colistin resistance (mcr-1 gene) in E. coli associated with colibacillosis in broiler chickens was found indicating a potential public health concern. Finally, the gene constellation of fimH, fimA, papC, iutA, and tsh was found to be the most prevalent virulence markers of APEC and could be used as fast pathotyping of avian E. coli isolates.
ACKNOWLEDGMENTS
This work was partially supported by the Faculty of Veterinary Medicine, Beni-Suef University, Egypt. The authors would like to thank D. Salama Shany, Lecturer of Poultry Diseases Department, Faculty of Veterinary Medicine, Beni-Suef University, Egypt for his help in molecular characterization of E. coli isolates.
Authors Contribution
Ali A; El-Sawah, AA: conceptualization. Ali, A; Abd El-Mawgoud, A; Dahshan, A; Ibrahim, A: methodology. Ali, A; Abd El-Mawgoud, A; Nasef, SA: Data analysis. Abd El-Mawgoud, A; Ali, A: writing-original draft preparation. All authors: reviewing and editing.
There is supplementary material associated with this article. Access the material online at:
Conflict of interest
The authors declare that there is no conflict of interest.
¥ The authors equally contributed to the manuscript
REFERENCES






