Advances in Animal and Veterinary Sciences
Research Article
Genetic Analysis of Infectious Bronchitis Virus Isolated from Egyptian Broiler Chicken Flocks between 2017 and 2018
Marwa Esmaiel1*, Gamilat Kotb2, Fatma Abdallah2, Ahmed A.H. Ali2
1Veterinary Administration of Afafos, Sharkia Governorate; 2Department of Virology, Faculty of Veterinary Medicine, Zagazig University, 44511, Zagazig, Egypt.
Abstract | Infectious bronchitis virus (IBV) is a major respiratory viral pathogen affecting chickens. In the current study, 81 tracheal tissues from 27 vaccinated broiler chicken flocks exhibited respiratory signs and high mortality rate up to 80% were collected from eight Egyptian Governorates during the period from 2017 to 2018. Out of the twenty-seven examined flocks, nine flocks were positive for IBV using quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR). Virus isolation was carried out for the RT-qPCR positive samples in embryonated chicken eggs (ECEs) for four blind passages along with partial amplification of spike protein (S1) using RT-PCR followed by sequencing for two representative isolates. Based on the deduced amino acids sequence and phylogenetic analysis of the S1 protein, the IBV isolates reported in this study were categorized into two lineages. One isolate, IBV-Ch-KafrAshaykh-2017 (MN201589) was belonged to GI-23 lineage showed high homology in amino acids sequences ranged from 92.4-99.4% with Egy/variant1 strains and the other isolate, IBV-Ch-Sharkia-2018 (MN201590) was belonged to GI-1 lineage showed high homology in amino acids sequences ranged from 95.3-98.8% with classical IBV strains. Therefore, it is very important to track the continuous evolution of IBV strains circulating in Egypt for better understanding the genetic relationship between circulating IBVs and the vaccine strains that will help in improving IBV control.
Keywords | Egypt, Broiler chickens, Infectious bronchitis virus, RT-PCR, Phylogenetic analysis
Received | September 11, 2019; Accepted | October 06, 2019; Published | October 10, 2019
*Correspondence | Marwa Esmaiel, Veterinary Administration of Afafos, Sharkia Governorate; Email: [email protected]
Citation | Esmaiel M, Kotb G, Abdallah F, Ali AAH (2019). Genetic analysis of Infectious Bronchitis Virus isolated from Egyptian broiler chicken flocks between 2017 and 2018. Adv. Anim. Vet. Sci. 7(s2): 57-62.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s2.57.62
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Esmaiel et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Infectious Bronchitis (IB) is a highly contagious respiratory viral disease affecting chickens and causes major economic losses to the poultry industry. It is characterized by severe respiratory distress in broilers while in layers it causes reduction in egg production (Ahmed and Ruja, 2006; Butcher et al., 2011). Infectious bronchitis virus (IBV) is a member of family Coronaviridae, genus Gammacoronavirus (Cavanagh, 2003). IBV has positive sense single stranded non-segmented RNA genome, about 27.6 kb in length. The virion has 4 structural proteins: nucleocapsid (N), membrane (M), envelope (E), and spike (S) (Jin-Ling et al., 2011). S protein is a major structure proteinof IBVwhich is sliced to 2 small protein subunits (S1 and S2) and it is the most variable gene in the IBV genome. The sequence of S1 gene can be used to differentiate IBVs into six IBV genotypes (GI-GVI), and 32 lineages (1-32) that have been identified worldwide (Valastro et al., 2016). S1 gene is about 1644 nucleotide in length and contains three hyper-variable regions (HVR 1, 2, and 3) that are responsible for induction of neutralizing and serotype specific antibodies (Haqshenas et al., 2005). Since 1930, many live attenuated IBV vaccines such as H120 and M41 have been introduced to counteract IBV infection (Sun et al., 2011). However, despite the intensive use of IBV vaccines, antigenically variable serotypes and newly emerged variants genotypes have affected the vaccinated flocks causing severe infection and vaccine failure (Jones, 2010). Changes of few amino acids in S1 subunit are able to generate newly genetic IBV variant strains (Adzhar et al., 1997). Therefore, genotyping of field strains is essential for investigation the new IBV variants in addition to assessthe currently applied vaccination regimes. The current study designed to study the epidemiology and evolution of IB viruses within the Egyptian broiler flocks during 2017 and 2018 and study the genetic variations between isolated field strains and their compatibility with different available commercially used vaccine strains.
MATERIALS AND METHODS
Ethical approval
The Animal Ethical Committee of the Faculty of Veterinary Medicine, Zagazig University, Egypt approved the design of the current study.
Sampling
Eighty-one samples from 27 broiler flocks of 24–35 days old suffered from respiratory signs and mortalities were collected during the period from February 2017 to January 2018. These samples were collected from eight Egyptian governorates (Sharkia, Qalyubiya, Dakahliya, North Sinai, Suez, Kafrelsheikh, Ismailia, and Giza). Briefly, pooled 3 tracheal tissues per each flockwere collected from freshly dead chickens. All samples were placed intovirus transport medium with 10% antibiotic solution containing 1000 IU penicillin, 1000 μg streptomycin, 2000 μg gentamycin (Pollard and Walker, 1997) and the clear supernatant fluid of each tissue homogenate was subjected to coolingcentrifugation at 3000 rpm /10 min and kept frozen at -20°C for further processing.
Detection of IBV using quantitative reverse transcriptase-polymerase chain reaction
Detection of IBV in the collected sampleswasconducted using quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) for detection of a 143-bp fragment of the 5´un-translated (5´-UTR) region of IBV (Callison et al., 2006). RNA was extracted from tissue homogenate supernatants using a QiaAmp viral RNA mini kit (Qiagen, Germany) following the manufacturer’s instructions. The extracted RNAswere reversed transcribed using Thermo ScientificTM Verso 1-step RT-PCR kit (Applied Biosystems, USA) according to the manufacturer’s instructions to produce cDNA. The amplification of the un-translated region of IBV was carried outusing forward IBV5_GU391, 5’-GCT TTT GAG CCT AGC GTT -3’and reverse IBV5_GL533, 5’-GCC ATG TTG TCA CTG TCT ATT G-3’, and probe IBV5_G, 5’-FAM-CACCACCAGAACCTGTCACCTC-BHQ1-3’ (Callison et al., 2006). RT-qPCR was conducted using Qiagen one step RT-qPCR Kit (Qiagen, Germany) following to the manufacturer’s instructions and Stratagene Mx3000P® QPCR System Real-Time PCR machine.
Isolation of IBV on embryonated chicken eggs (ECEs)
The supernatant of RT-qPCR positive samples (0.2 ml) was inoculated into the allantoic sac of 11-day-old ECEs (five ECEs for each sample) from commercial non-vaccinated chickens. After 48-96 hours, the allantoic fluids of inoculated eggs were harvested and pooled. For each sample, four successive blind serial passages were conducted. The inoculated eggs in the last passage were left in egg incubator for 7 days at 37°C with daily examination to see the pathognomonic characteristic changes of embryos (curling, stunting, and dwarfing) (Delaplane, 1947) and the allantoic fluids were harvested and kept at _70 °C.
Amplification of spike 1(S1) gene using conventional RT-PCR
S1 gene of nine positive isolates were partialamplified using conventional RT-PCR using Qiagen one step RT-PCR Kit (Qiagen, Germany) and forward primer IBV-S1-F 5’-CACTGGTAATTTTTCAGATGG-3’and reverse primer IBV-S1-R 5’-CAGATTGCTTACAACCAC C-3’ (Adzhar et al., 1997). The ampliconswere purified using the QIA quick gel extraction kit (Qiagen, Germany) following the manufacturer’s instructions, and kept at -20°C until sequencing.
Phylogenetic analysis
The amplified S1 gene of two selected isolates was sequenced in both directions using Big dye Terminator V3.1 cycle sequencing Kit (Perkin-Elmer, CA) utilizing the amplification primers in applied Biosystems 3130 genetic analyzer (ABI, USA). The nucleotide sequences of S1 gene of the twoisolates (IBV-Ch-Kafr Ashaykh-2017and IBV-Ch-Sharkia-2018) were submitted to the GenBank database under the accession numbers MN201589 and MN201590, respectively. The nucleotide and aminoacid sequences of the two isolates were compared and aligned with the sequences of representative strains of different IBV genotypes existing in GenBank (Table 1). Comparativeamino acid alignment was carried out using Clustal W Multiple alignment of Bio Edit Version 7.0 software (Hall, 1999). Amino acid sequence identities and divergences were calculated utilizing Meg Align software (DNA STAR® Laser gene® version 7.2, USA). Phylogenetic tree was created via the maximum-likehood method employing the Kimura 2-parameter model in MEGA6.06 software (Tamura et al., 2013) with boots trapping 1000.
Table 1: Sequences of reference IBV strains published in GenBank which used in this study for the phylogenetic analysis.
| GenBank Accession Number | Reference IBV Strains | Country of Isolation |
| JX173489 |
Eg/CLEVB-1/IBV/012 |
Egypt |
| EU780077 |
IS/1494/06 |
Israel |
| KU238171 |
D1344/2/4/10_EG |
Egypt |
| JX174183 | Egypt/Beni-Suef/01 | Egypt |
| JX569792 | ck/CH/HN/1205 | China |
| EU350550 | IS/1366 | Israel |
| GU393335 | H120 | Netherlands |
| AY279533 | IS/885 | Israel |
| AY091552 |
Israel/720/99 |
Israel |
| KU979006 |
IBV-EG/1442F-SP1-2014 |
Egypt |
| JX174186 |
Ck/Eg/BSU-3/2011 |
Egypt |
| KC197201 |
IBV/ck/Egypt/12vir6109-4/2012 |
Egypt |
| KC608180 |
EGY/12773F-3 |
Egypt |
| JQ839289 | Eg/11539F | Egypt |
| MH745418 | IB-54NB-chicken-LEBNAN | Lebanon |
| MK562092 | IBV/Ck/EG/Fadllah-10/2019 | Egypt |
| DQ487085 | Egypt/F/03 | Egypt |
| KM067900 | CR88-UPM2013 | Malaysia |
| AF093796 | variant 2 | Israel |
| MK310099 | Sharkia/2013 | Egypt |
| AY135205 | IS/236 | Israel |
| MH745419 | IBV_Duck_50NL | Lebanon |
| AY561713 | MA5 | USA |
| KF377577 | 4/91 | China |
| AY561711 | M41 | USA |
RESULTS
Detection of IBV in field samples using qRT-PCR
Out of 27 tested flocks, nine flocks were positive for IBV using RT-qPCR which were geographically distributed as follow; five positiveflocks out of 14 flocks from Sharkia governorate, one out of four flocks from Dakahliya and oneflockfrom the following governorates (Qalyubiya, Kafrelsheikh and Giza). However, Ismailia, Suez and North Sinai were negative (Table 2).
Isolation of IBVon ECEs and detection of S1 gene of IBV using conventional RT-PCR
Nine isolates, which were positive by RT-qPCR, were subjected for virus isolation and clear pathognomic lesions; curling, stunting, and dwarfing were observed after four blind passages (Figure 1). Conventional RT-PCR was conducted on the collected allantoic fluid for nine isolates that revealed specific amplification at the expected size ofpartial S1 gene (400 bp) (Figure 2). Then, PCR products of two isolates were chosen for sequencing.
Table 2: Results of IBV detection using qRT-PCR.
| Governorate | Number of tested flocks | Vaccination status of flocks | Number of positive flocks |
| Sharkia | 14 |
MA5+ H120 + 4/91 |
5 |
| Qalyubiya | 1 | MA5+ H120 | 1 |
| Suez | 2 | MA5+ H120 | 0 |
| North Sinai | 2 | MA5+ H120 | 0 |
| Kafrelsheikh | 1 |
MA5+ H120 + 4/91 |
1 |
| Giza | 1 | MA5+ H120 | 1 |
| Dakahliya | 4 | MA5+ H120 | 1 |
| Ismailia | 2 | MA5+ H120 |
|
Amino acid sequence identity and phylogenetic analysis
Two representative isolates (IBV-Ch-Kafr Ashaykh-2017 and IBV-Ch-Sharkia-2018) were selected forgenetic analysis of S1 gene. Briefly, the two-sequenced isolates have amino acid identity 77.4% compared to each other. IBV-Ch-KafrAshaykh-2017 (MN201589) has amino acid identity ranged from 92.4- 99.4 % to group (Egy/variant 1) of GI-23 lineage as Sharkia/2013, IBV_Duck_50NL, IB-54NB-chicken-LEBNAN, variant 2, Eg/CLEVB-1/IBV/012, IS/1494/06, Eg/CLEVB-1/IBV/012, IBV-EG/1442F-SP1-2014 and D1344/2/4/10_EG strains. Whilst, IBV-Ch-Sharkia-2018 isolate under accession number MN201590 showed amino acid identity ranged from 95.3-98.8 % to classical genotype of GI.1 lineage as MA5, H120, M41, IS/236 and Egypt/F/03 strains. On the other hand, IBV-Ch-KafrAshaykh-2017 and IBV-Ch-Sharkia-2018 have amino acid identity percentage (76.1 and 71.4%) with 4/91 genotypes, respectively (data not shown).
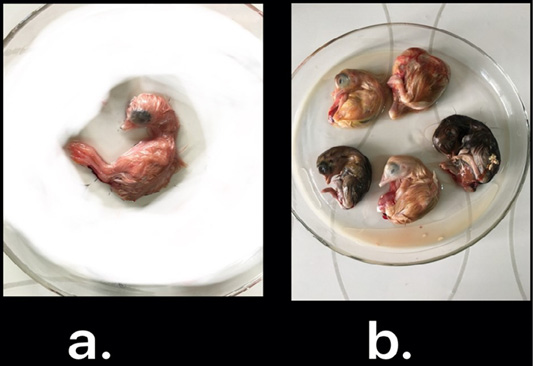
Figure 1: IBV isolated in ECEs. Normal embryo development (a) and curled, stunted, and dwarfed embryos infected with IBV after fourth passage (b)
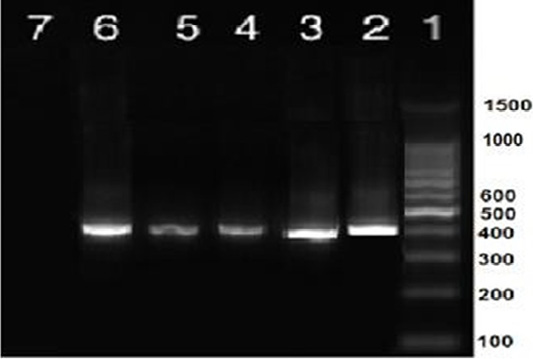
Figure 2: RT-PCR amplification of partial S1 gene from IBV isolates in the present study showed a single specific band (400bp). Lane 1: 1 kbp DNA ladder (Fermantas); Lane 2, positive control; Lanes 3–6: positive IBV-infected samples. Lane 7: negative control.
The deduced amino acidsequences of HVR1 and HVR2 spanning the region from 60-88 and 115-140, respectively in the S1 protein for the two isolates reported in this study were compared with classicalgenotype of GI-1 lineage (H120, MA5, M41) and 4/91 genotype of GI-13 lineage (4/91). IBV-Ch-KafrAshaykh-2017 showed distinct patterns in HVR1 and HVR2 of Sp1 protein compared to the H120, MA5, M41 and 4/91strains as follows at HVR1; T69S, F70I, Y71H, E72W, Y74K, I76F, A79S. While at HVR2, the substitutions were F115Y, S117N, Q118G, N131D as shown in Figure 3.

Figure 3: Comparison between deduced amino acid sequences of HVR1 (amino acid position from 60-88) and HVR2 (amino acid position from 115-140) of S1 protein of two isolates reported in the study and currently used vaccine strains in Egypt.
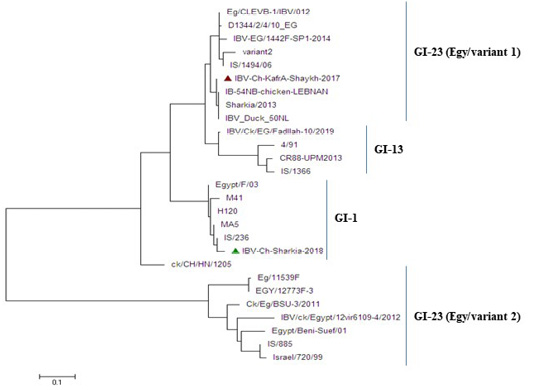
Figure 4: Phylogenetic tree based on nucleotide sequences of S1 gene for the two IBV isolates and related 25 reference IBV strains. The IBV-Ch-KafrAshaykh-2017 isolate marked with a red triangle belonging to isolates of GI-23 lineage (Egy/variant1 group), while IBV Ch-Sharkia-2018 marked with a green triangle belonging to isolates of GI-1lineage (classical group).
The phylogenetic analysis revealed that the two IBV strains were clustered into two distinctgroups: group (Egy/variant 1) of GI-23 lineage and classical genotype ofGI.1 lineageas shown in Figure 4. Whereas, IBV-Ch-KafrAshaykh-2017 isolate under accession number MN201589 was closely related to group (Egy/variant 1) of GI-23 lineage of IS/1494/06 genotype origin and very close to previously isolated Egyptian IBVs (Sharkia/2013, Eg/CLEVB-1/IBV/012, Eg/CLEVB-1/IBV/012,IBV-EG/1442F-SP1-2014 and D1344/2/4/10_EG). However, IBV Ch-Sharkia-2018 isolate under accession number MN201590was clustered together with classicalstrains (Egypt/F/03, IS/236, H120, MA5and M41) in GI.1 lineage.
Discussion
IBV has been reported since 1930s (Schalk and Hawn, 1931); however, IB outbreaks are continuously reported repeatedly (Zanatyet al., 2016; Feng et al., 2017; Lounas et al., 2018; Rohaim et al., 2019). In the present study, the prevalence of IBV infection in 27 broiler chicken flocks was investigated by RT-qPCR that revealed that nine flocks were positive for IBV.
The nine RT-qPCR confirmed samples were isolated through inoculated into allantoic sac of 11-day-old ECEsthat showed curled, stunted, and dwarfed embryos after fourpassages (Figure 1). Five ECEs of the 4th passage were inoculated with Egypt/F/03 and incubated till being 18-day-old and the embryos of 5 eggs exhibited standard IBVpathognomonic embryo changes (stunting, curling and dwarfing) (Abdel-Moneim et al., 2006).
Sequencing of HVRs of S1 gene is significant for IBV genotyping. In the present study, sequence analysis of HVR1 and HVR2 of S1 protein of thetwo isolates showed that the amino acid identity percent was 77.4% between each other. The amino acid similarity between IBV-Ch-KafrAshaykh-2017 and other strains isolated from the bordering countries like Israel was ranged from 92.9% to 99.4%. This comparison is significant as a result of uncontrolled movement of animal populations and trafficking across country borders (Bochkov et al., 2006).
Currently, MA5, M41, H120 and 4/91 IBV vaccines have been applied in the face of IBV infections in Egypt. Although the use of these vaccines, newly emerged variant IBV strains still continuously evolving (Zanaty et al., 2016; Rohaim et al., 2109). In this study, the phylogenetic analysis classified two isolates into two different lineages, whereas IBV-Ch-Kafr Ashaykh-2017 isolate was located in group (Egy/variant 1) of GI-23 lineage and IBV-Ch-Sharkia-2018 isolate was located in classical genotype of GI.1 lineage as shown in (Figure 4). In the present study, Figure 3 explained that HVR1 and HVR2 had amino acid substitutions where isolate that belong to group (Egy/variant 1) of GI-23 lineage had amino acid substitution at position T69S in receptor binding site of the variant strains. T69 have been defined to be important for binding of the IBV spike protein to the chicken respiratory tract (De Witt, 2000). In addition to, small change in the amino acid sequence of S1 protein can result in generation of novel genotypes that differ antigenically from the existing classic and variant vaccine strains (Abozeid et al., 2017) as well as, changes as little as 5% in the S1 gene are able to alter the protective ability of IBV vaccines (Wang and Huang, 2000). Our results explained that IBV-Ch-KafrAshaykh-2017 had a distant relation to commercially used vaccine strains (H120, M41, MA5 and 4/91) in Egypt. These results suggest that the applied vaccination regimen was inadequate to provide protection, or chickens are exposed to other respiratory pathogens by stress during vaccination, which decreases immunity (Yilmaz et al., 2016).
Conclusion
Our results revealed that the isolated IBVs in the current study from Egypt during 2017-2018 are categorized into two groups. IBV-Ch-KafrAshaykh-2017 isolate clustered with group (Egy/variant 1) of GI-23 lineage of IS/1494/06 genotype origin and IBV-Ch-Sharkia-2018 isolate was more closely related to IB vaccine strains like Mass-type (H120, M41 and MA5) that point to independent IBV evolution and persistence of divergent IBV strains distributing in Egypt. Therefore, it is important tocarry out more epidemiology studies to better understand the spreading nature IBVs in Egypt and study the genetic relationship between circulating IBV field strain and vaccine strains in order to improve IBV control strategies.
ACKNOWLEDGEMENTS
This work was supported by Faculty of Veterinary Medicine, Zagazig University, Egypt.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
AUTHORS CONTRIBUTION
All authors contributed equally in this work.
REFERENCES






