Advances in Animal and Veterinary Sciences
Research Article
Molecular Characterization and Phylogenetic Analysis of Fasciola gigantica in Iraqi Sheep Using ITS1
Reedha N. Hamoo1*, Fouad S.I. Al-Rubaye1, Nashaat G. Mustafa2
1Department of Biology, College of Education for Girls, University of Mosul, Mosul, Iraq; 2Department of Physiology, Biochemistry, and Pharmacology, College of Veterinary Medicine, University of Mosul, Mosul, Iraq.
Abstract | Fasciolosis is a worldwide disease of man and animals result in huge economic losses. The objective of this study was to characterize for the first time Fasciola gigantica in Iraq employing molecular approaches. In this work, 65 adult fasciola worms were isolated from sheep liver of different age and both sex during May-August 2017, in Kirkuk city, Iraq. Molecular analysis conducted on the internal transcribed spacer 1 to characterize the species of parasite depending on the sequence analysis. Results reveal that the isolated worms were Fasciola gigantica and they have a great similarity, up to 100%, compared with other global isolates. In addition, results documented a single nucleotide polymorphism, at the 276th nucleotide of the studied fragment, and the isolates of our project highly related to various global isolates by using phylogenetic tree analysis. In conclusion,our isolates are faithfully analogous to global isolates with common SNP at 276th nucleotide, and ITS1 gene considers a novel marker in identifying Fasciola gigantica and study the relation with other isolates.
Keywords | Fasciola gigantica, Internal transcribed spacer 1 gene, Iraqi sheep, Phylogenetic tree.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | November 26, 2018; Accepted | December 18, 2018; Published | January 18, 2019
*Correspondence | Reedha N. Hamoo, Department of Biology, College of Education for Girls, University of Mosul, Mosul, Iraq; Email: [email protected]
Citation | Hamoo RN, Al-Rubaye FSI, Mustafa NG (2019).Molecular characterization and phylogenetic analysis of fasciola gigantica in iraqi sheep using ITS1. Adv. Anim. Vet. Sci. 7(4): 256-260.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.4.256.260
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Hamoo et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Fasciolosis is a prevalent liver fluke disease of ruminants, particularly sheep and cattle caused by a trematode Fasciola hepatica (F. hepatica) and/or Fasciola gigantica (F. gigantica), and it is still a big problem issue due to the giant global economic losses of about 2 million dollars annually (Spithill et al., 1999). The disease was reported worldwide, particularly in certain areas of South America, Egypt, and Iran in long list mammalian species including human (O’Neill et al., 1998; Rokni et al., 2002). Nearby 2.4 million of the man infected by fasciola around the world, and millions still living at the high risk of infection (Mas-coma et al., 1999). The infection may result from F. hepatica, or F. gigantica, or mixed infection of both. Interestingly, hybridization of both species may occur inside host body, and the subsequent offspring have intermediate phenotypes with different polidies; either diploid, or triploid, or mixoplid (Vara-Del Rio, 2007; Beesley et al., 2017).
Various molecular methods were employed in the diagnosis, taxonomy, and the study of fasciola like whole genome sequencing (WGS) and single nucleotide polymorphism (SNP) (Cwiklinski, 2015), Random amplified polymorphic DNA (RAPD) (Semyenova, 2003; Gunasekar et al., 2008), and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) (Walker et al., 2007; Simsek et al., 2011). Internal transcribed spacer (ITS) located between 5.8S and 28.5S of nuclear ribosomal DNA, was utilized in genetic characterization and polymorphism of fasciola worldwide since ITS has many various fragments with highly conserved sequence (Morgan and Blair, 1995; Choe et al., 2011). Authors suggested that ITS sequence analysis of fasciola from different global regions revealed only a few (3-6) nucleotide polymorphism (Choe et al., 2011).
However, Inour country only a few studies concerned with F. gigantica distribution in intermediate and final hosts (Farage, 1998; Al-Mayah, 2004; Gzi and Rahma, 2012; Shahatha, 2013), but the molecular study of F. gigantica has never been done in Iraq until now. Therefore the aim of this project was to analyze ITS sequences and phylogenetic tree of F. gigantica isolated from the liver of sheep in Kirkuk city in Iraq.
Materials and methods
Parasite Samples
Adult 65 fasciola worms were collected from the liver of sheep through inspection process in the slaughterhouse of the central abattoir of Kirkuk city, Iraq, at May-August 2017. Fasciola gigantica were identified by their characteristic morphology, and the isolated worms were gently washed three times by phosphate buffer saline (PBS), fixed in 70% ethanol and stored at room temperature until DNA extraction.
DNA Extraction
About 15 mg of a lateral zone of adult fasciola worms were removed and crashed between two glass microscopic slides, the ethanol in each sample was permitted to evaporate for a few minutes and washed with distilled water for many times. DNA extracted according to manufacturer guide using G-spin DNA extraction kit (Cat No.17045/Intron Biotechnology/Korea), and the purity of DNA was evaluated by Nanodrop (2000/2000c spectrophotometer, Thermo Scientific/USA). Extracted DNA stored at -20°C until used in the PCR amplification.
Primer Design
The primers were designed to amplify 498 bp fragment of IST1 gene using forward and reverse primer (Integrated DNA technologies, USA) (Table 1). The primers dissolved in the free dH2O to give a final concentration of 100 pmol/µl as stock solution and kept a stock at -20 to prepare 10 pmol/µl concentration as work primer suspended, 10 µl of the stock solution added to 90 µl of the free dH2O water to reach a final volume 100 µl.
Table 1: The primers were used in amplification of ITS1.
| Primer | Sequence | Tm (ᵒC) | GC (%) | Product size |
| Forward | 5'- ACCGGTGCTGAGAAGACG- 3' | 57.3 | 61.1 |
498 base pair |
| Reverse | 5'- CGACGTACGTGCAGTCCA- 3’ | 57.4 | 61.1 |
PCR
PCR PreMix kit (i-Taq, 25025 Intron, Korea) reaction composed of 5 µl Taq PCR PreMix [i-Taq DNA Polymerase (5 U/μl), dNTPs (2.5 mM), reaction buffer (10 X) (1 X), gel loading buffer (1 X)], 10 picomols/µl F primer and 10 picomols/µl R primer, 1.5 µl of extracted DNA (1.5 µl of ddH2O instead of DNA samples were used as a negative control), 16.5 µl of ddH2O, the final volume of PCR reaction was 25µl. the mixture amplified by thermal cycler (MultiGeneTM OptiMax Gradient, Labnet, USA), under the following conditions: initial denaturation 94˚C for 5 min, followed by 30 cycles at 94˚C for 30 sec (denaturation), 55˚C for 30 sec (annealing), 72˚C for 2 min (extension), and 72˚C for 10 min (final extension).
Gel Electrophoresis
The PCR products were separated on a 2% agarose gel electrophoresis (CBS, Scientific, USA), at 5 volt/cm2. 1x TBE buffer (IBS.BT004, Conda, USA) for 1.30 hours, and visualized by exposure to ultra-violate light (302 nm) illuminator (Vilber lourmat, France) after ethidium bromide (Intron, Korea) staining.
Sequencing and Sequence Analysis
Five F. gigantica samples from sheep were sequenced using 15 µl of PCR product and F and R primers. The sequencing was done depending on national instrumentation center for environmental management (nicem) online at (http://nicem.snu.ac.kr/main/?en_skin=index.html), biotechnology lab machine is DNA sequencer (3730XL, Applied Biosystem). The homology search was conducted using Basic Local Alignment Search Tool (BLAST) program which is available at the National Center for Biotechnology Information (NCBI) online at (http:// www.ncbi.nlm.nih.gov) and BioEdit program.
Results
In this study, 65 isolates of adult F. gigantica were investigated from 28 naturally infected sheep in the central abattoir of Kirkuk city, Iraq. Genomic DNA was productively extracted and the specific primers (Table 1) were used in this work successfully amplified a fragment of 498 bp of a part of ITS1 gene of fasciola samples, while negative control produces no bands on the gel (Figure 1).
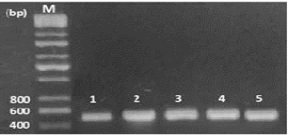
Figure 1: Electrophoresis of PCR product of ITS1, the band size (Lane 1-5 F. gigantica) 498 bp. Lane M: DNA ladder (200 bp).
Table 2: SNP of F. gigantica ITS1 region from sheep liver in Iraq.
| No. Of sample | Type of substitution | Location | Nucleotide | Range of nucleotide | Sequence ID | Score | Expect | Identities |
Source
|
| 1 | Transition | 276 | G>A | 126 to 508 |
ID: MF372919.1 |
702 | 0.0 | 99% | Fasciola gigantica |
|
3 |
Transition | 276 | G>A | 127 to 529 |
ID: MF372919.1 |
739 | 0.0 | 99% | Fasciola gigantica |
| 4 |
|
|
-------- | 126 to 529 |
ID: MF372919.1 |
747 | 0.0 | 100% | Fasciola gigantica |
| 5 | Transition | 247 | G>N | 126 to 529 |
ID: MF372919.1 |
737 | 0.0 | 99% | Fasciola gigantica |
| Transition | 276 | G>A |
Alignment analysis through online blast reveals similarity of 100 % (sample 4) and 99 % (samples 1, 3, and 5) (Table 2), of inspected sequence fragment when compared with Genbank database of F. gigantica available reference data. Furthermore, it’s clear and interesting that 3 of 4 (75 %) of samples expose single nucleotide polymorphism (G to A) of the 276th nucleotide of an investigated fragment, while one sample (25 %) shows SNP (G to N) of 247th nucleotide (Table 2 and Figure 2). One of the interesting novel result of this paper, our sequence of IST of F. gigantica deposited (Accession No. MG786553, Ver. MG786553.1) in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/1328159525).
The phylogenetic tree was formed according to Iraqi isolate of F. gigantica compared with most identical international isolates, our results demonstrate that the most similarity to KC476171.1 Bangladesh Isolates of F. gigantica followed by other isolates (Figure 2).

Figure 2: Phylogenetic relationship with global isolates based on ITS1 sequence of F. gigantica isolated from the liver of sheep in Iraq.
Discussion
In addition to precise consequences of molecular biology approaches in the study of parasites and their genes, this field grows quickly and extends widely over the world, promising the goal achievement of their employment. As mentioned earlier, this is the first study in Iraq deals with the molecular investigation of F. gigantica ITS gene. The high incidence of F. gigantica infection agree with results of other investigations (by ordinary serological and morphological methods) in the middle and south, but not north of Iraq, like Farage (1998), Al-Mayah (2004),Gzi and Rahma (2012).
Alignment analysis confirmed our results and reveal extreme similarity with other isolates, and this outcome may be due to that our isolate is relatively recently introduced to Iraq. Authors believe that F. gigantica diverged from F. hepatica 17 million years ago (Irving et al., 2003) then F. gigantica spread in Asia, Middle East, and the Far East up to become one of the harmful parasitic infection of cattle and sheep (MacManus and Dalton, 2006; Raina et al., 2015; Bazh et al., 2016). Beside ITS sequence analysis were utilized to discrimination of fasciola species (Huang et al., 2004; Peng et al., 2009), the kind, number, and location of SNP of this region can be considered a critical marker in matching between different isolates. Our findings are in a good agreement with other authors (Huang et al., 2004; Prasad et al., 2008) whose shown that SNP of ITS can be concerned with the 276th nucleotide of ITS1 region. The sequence of ITS1 with SNP may be due to the presence of hybrids in our isolates, as mentioned by Ichikawa-Seki (2017). The main limitation of our experimental results is that it is restricted to Fasciola gigantica isolated from sheep liver only in Kirkuk city in Iraq. From the outcome of investigations it is possible to conclude that ITS1 region is a novel part for molecular characterization and phylogenetic study of Fasciola gigantica, also our isolates are closely similar to global isolates with common SNP at 276th nucleotide.
Acknowledgements
Authors would like to thanks College of Education for Girls, University of Mosul, Mosul, Iraq, and central abattoir of Kirkuk city, Iraq, for their nonfinancial support of this work.
Conflict of interest
The authors declare that they have no competing interests, financial or otherwise.
authors contribution
Reedha N Hamoo designed the experiments and analysed the data. Fouad S.I. Al-Rubaye collected samples and analysed the data and Nashaat G. Mustafa carried out the experiments and wrote the manuscript with input from all authors.
References






