Advances in Animal and Veterinary Sciences
Research Article
Molecular Characterization of Bluetongue Virus Serotype 16 from Andhra Pradesh, India
Prasad Minakshi1*, Koushlesh Ranjan2, Gaya Prasad3
1Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar, Haryana, 125004; 2Department of Veterinary Physiology and Biochemistry, Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut, Uttar Pradesh, 250110; 3Indian Council of Agricultural Research (ICAR), New Delhi, India.
Abstract | Bluetongue (BT) is a Culicoides borne infectious disease of domestic and wild ruminants. In the present study, a total of four BTV samples (GNT27/IND, MBN48/IND, MBN50/IND and VJW66/IND) of sheep origin from Andhra Pradesh state of India were inoculated to 9-11 days old chicken embryos. The BTV samples showed specific cytopathic effect (CPE) in BHK-21 cell culture. The samples also showed BTV specific characteristic migration pattern of 3:3:3:1 in RNA-Polyacrylamide gel electrophoresis. Furthermore, the ns1 gene based group specific RT-PCR confirmed the samples to be BTV. Further, serotyping based on vp2 gene confirmed these samples as of BTV16 serotype. The sequence analysis of vp2 gene specific PCR products (768bp) revealed a high degree of (91-99% nucleotide and amino acid) identity within BTV isolates of this study and other BTV16 isolates from India. Further sequence analysis revealed highest nucleotide (87.7-99%) and deduced amino acid (91.3-99.5%) sequence identity with Eastern viruses from India, Japan, China, South Africa, Italy and Israel. However, it showed only 71.0-75.9% nucleotide and 81.1-87.6% deduced amino acid sequence identity with western BTV16 from Nigeria, confirmed isolates of this study as eastern topotype. The phylogenetic analysis showed a close clustering between isolates of this study and other Indian BTV16 isolates which were closely related to Japan, China and Greece isolates. The molecular analysis revealed that the isolates in the study are very close to Greece, China, Japan and other Indian isolates of BTV16.
Keywords | Bluetongue virus 16, Topotype, NS1 gene, VP2 gene, RT-PCR, Sequencing, phylogenetic analysis
Editor | Sandip Kumar Khurana, Principal Scientist, National Center on Equines (NRCE), Sirsa Road, Hisar, Haryana, 125004, India.
Special Issue | 1(2015) “Biotechnological and molecular approaches for diagnosis, prevention and control of infectious diseases of animals”
Received | March 15, 2015; Revised | April 07, 2015; Accepted | April 08, 2015; Published | April 21, 2015
*Correspondence | Minakshi Prasad, LLR University of Veterinary and animal Sciences, Hisar, India; Email: [email protected]
Citation | Minakshi P, Ranjan K, Prasad G (2015). Molecular characterization of bluetongue virus serotype 16 from Andhra Pradesh, India. Adv. Anim. Vet. Sci. 3(1s): 22-27.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.1s.22.27
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Minakshi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bluetongue (BT) is an infectious but non-contagious viral disease of domestic and wild ruminants. It is transmitted by Culicoides vector (MacLachlan, 1994) and caused by Bluetongue virus (BTV) which belongs to genus Orbivirus under family Reoviridae. BT causes severe economic loss to livestock industry due to its high morbidity, mortality, foetal abnormality, still birth, abortion, wool break, weight loss, reduced meat and milk yield. The severe form of the disease is seen in sheep and white-tailed deer (Howerth et al., 1988; Darpel et al., 2007). Other domestic ruminants such as cattle, buffalo and goats act as silent reservoirs and remain viraemic for several months post infection (MacLachlan et al., 2009). The clinical form of disease is characterized by pyrexia, coronitis, swelling of tongue and lips, cyanotic discoloration of tongue and muzzle leading to death. The subclinical infection of BTV can also cause reduced milk yield, loss of condition and abortion leading to infertility in animals (Osburn, 1994). Due to severe economic losses mandatory trade barrier and surveillance have been imposed on movement of ruminant animals, their germplasm and other animal products from BT endemic countries to BT free countries (Velthuis et al., 2009). Since, BT can infect several species of animals, therefore now it is listed as multi species disease by Office International des Epizooties (OIE, 2013).
The genome of BTV particle consists of ten segmented linear double-stranded RNA (dsRNA). The BT genome segments encode 7 structural proteins (VP1 to VP7) essential for viral nucleic acid replication and viral capsid assembly. They also encode 4 non-structural proteins (NS1, NS2, NS3/ NS3a and NS4) which are produced in infected host cell and have role in egression of viral particle from cell (Ratinier et al., 2011). The inner capsid of virion particle is composed of two major proteins (VP3 and VP7) and three minor proteins (VP1, VP4, and VP6). The outer capsid is composed of VP2 and VP5 proteins which are major and minor serotype specific for individual BTV serotype.
The segmented nature of BTV genome facilitates reassortment in genome segments with other BTV which lead to evolution of new serotypes. Twenty-four distinct BTV serotypes (BTV1 to BTV24) have been reported worldwide (Mertens et al., 2004). However, two new serotypes, BTV25 and BTV26 from Switzerland and Kuwait, respectively (Hofmann et al., 2008; Maan et al., 2011) have also been reported. Since, India is endemic for Culicoides vector, a large no of BTV serotypes have been reported from India. Serum neutralization and virus isolation in cell culture have revealed the prevalence of 22 different BTV serotypes in Indian subcontinent (Prasad et al., 2009; Susmitha et al., 2012). Several serotypes of BTV have been isolated from Andhra Pradesh. In this study BTV isolates of sheep origin from Andhra Pradesh state were characterized using vp2 gene specific RT-PCR followed by nucleic acid sequencing and phylogenetic analysis with similar serotypes from India and abroad.
Materials and methods
Sample Preparation
The blood sample was collected from Nellore breed of sheep from different geographical region of Andhra Pradesh state in 2009. All the sheep were showing symptoms of BT such as pyrexia (103-104oF), lameness, swelling of muzzle, salivation, cyanosis of lip and tongue. A total of four samples from Guntoor (isolate, GNT27/IND), Mahboobnagar (isolate MBN48/IND and MBN50/IND) and Vijayawada (isolate VJW66/IND) region were collected. The blood samples were processed and inoculated to 9-11 day old chicken embryo through intravenous route. After showing embryopathic effect the embryos were harvested within 7 days and inoculated to day old monolayer of BHK-21 cell culture.
Nucleic Acid Extraction and RNA-PAGE
After appearance of about 75% cytopathic effect (CPE) in BHK-21 cell culture, the cells were harvested and centrifuged at 2000Xg for 10 minutes (Remi, India). The viral nucleic acid from all the four samples was extracted from pelleted cell culture material using Guanidinium isothiocynate method (Chomoczynski and Sacchi, 1987). The viral nucleic acid (dsRNA) of all the four samples was subjected to 8% poly acrylamide gel electrophoresis followed by silver staining to visualize the BTV specific migration pattern of nucleic acid (Svensson et al., 1986).
cDNA preparation and confirmation of BT isolates
The viral nucleic acid of all the four samples was used for cDNA preparation using random decamer primer (Ambion, USA) and moloney murine leukemia (Mo-MuLV-RT) virus reverse transcriptase enzyme (Sibzyme, Russia). The reaction mixture containing 7µg of viral dsRNA, 6% DMSO and 30 pMol of random decamer primer was allowed for heat denaturation. The final volume of reaction mixture was made to 50µl using 400µM each dNTPs and 500 U of Mo-MuLV reverse transcriptase enzyme. The decamer primer was allowed to anneal at 25°C for 10 minute followed by reverse transcription at 37°C for 60 minute in thermal cycler (Biorad i-Cycler, USA). To confirm the samples as BTV, the cDNA thus obtained were allowed to group specific ns1 gene based PCR using primer pairs F: 5’GTTCTCTAGTTGGCAACCACC3’ and R: 5’ AAGCCAGACTGTTTCCCGAT3’ to produce an amplicon of 274bp size (Prasad et al., 1999).
Determination of BTV Serotype and Nucleic acid Sequencing
The serotypes of all the virus isolates were confirmed by vp2 gene serotype specific RT-PCR using primers specific to all 24 serotypes. The cDNA were subjected to PCR using all the serotype specific primers individually in a 20 µl reaction mixture 2 µl cDNA, 20 µM of serotype specific primers, 3% DMSO, 0.4 µl of 10mM dNTPs mix (Finnzyme, Finland), 4 µl 5X HF buffer and 0.4 U (2U/ µl) phusion high- fidelity DNA polymerase (Finnzyme, Finland) in a thermal cycler (Biorad iCycler, USA). The PCR amplification programme was set as initial denaturation for 2 minute at 98°C, followed by 35 cycles of denaturation at 98°C for 10 second, primer extension at 72°C for 20 second and annealing at 55°C for 20 second. The final extension was allowed for 10 minute at 72°C. The PCR products were allowed to 1% agarose gel (Sigma, USA) electrophoresis and were visualized using gel documentation system (Biovis, USA). The PCR products were purified using QIA quick gel extraction kit (Qiagen, USA). The final serotype confirmation was done by nucleic acid sequencing of purified PCR products using serotype specific forward and reverse primers. The sequencing reaction was performed using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) in Genetic Analyser ABI PREISM TM 3130 XL machine as per the manufacturer’s instruction in our laboratory.
Nucleotide Sequence Analysis
The nucleic acid sequence based serotype confirmation was done using online available BLASTN+ 2.2.31 search (Zhang et al., 2000). The forward and reverse sequences of each isolate were aligned to generate contig sequences using Bioedit v7.2.3 software (Hall, 1999). The contig sequences were used for further analysis. The nucleotide as well as deduced amino acid percent identity with global isolates of BTV16 was calculated using Bioedit v7.2.3 software (Hall, 1999). The phylogenetic analysis of nucleotide sequences of all the isolates along with other global sequences were done using Mega 6 software programme (Tamura et al., 2013).
Results and discussion
India is a tropical country with typical monsoon climate which provides suitable environment for Culicoides vector multiplication. Out of more than 1400 different Culicoides species reported worldwide about 63 are morphologically identified from different geographical regions of India (Halder et al., 2013; Archana et al., 2014). The large numbers of Culicoides vectors explain the prevalence of 22 different BTV serotype in India. The antibodies against BTV have been detected in several species of domestic and wild ruminants in India (Prasad et al., 1998). In the present study, four BTV isolates of sheep origin adapted in BHK21 cell line were used for serotyping and molecular characterization.
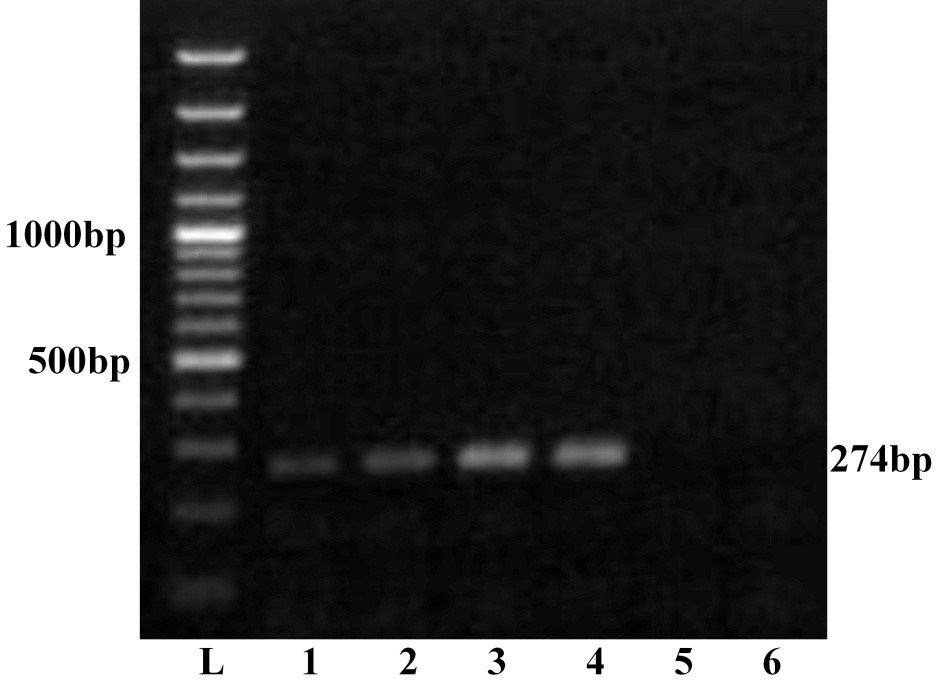
The four viral isolates viz. GNT27/IND, MBN48/IND, MBN50/IND and VJW66/IND were adapted to BHK-21 cell lines and were passaged up to tenth passage for bulk production of virus particle. All these isolates produced BTV specific CPE such as rounding and aggregation of cells, vacuolation in cells, floating of dead cells in medium within 36 hours. The viral nucleic acid extracted from pelleted cell culture materials of all the isolates was screened by RNA-PAGE and silver staining. The RNA-PAGE analysis revealed BTV specific 10 dsRNA segments arranged in 3:3:3:1 pattern (data not shown) in all the samples. The viral nucleic acid was allowed for ns1 gene group specific RT-PCR. Agarose gel electrophoresis of ns1 gene RT-PCR product showed 274 bp of expected amplicon size in all the samples, confirming the samples as BTV (Figure 1). Thus characteristic CPE in BHK-21 cell culture, specific migration pattern of viral nucleic acid in RNA-PAGE and ns1 gene group specific RT-PCR confirmed the samples as BTV.
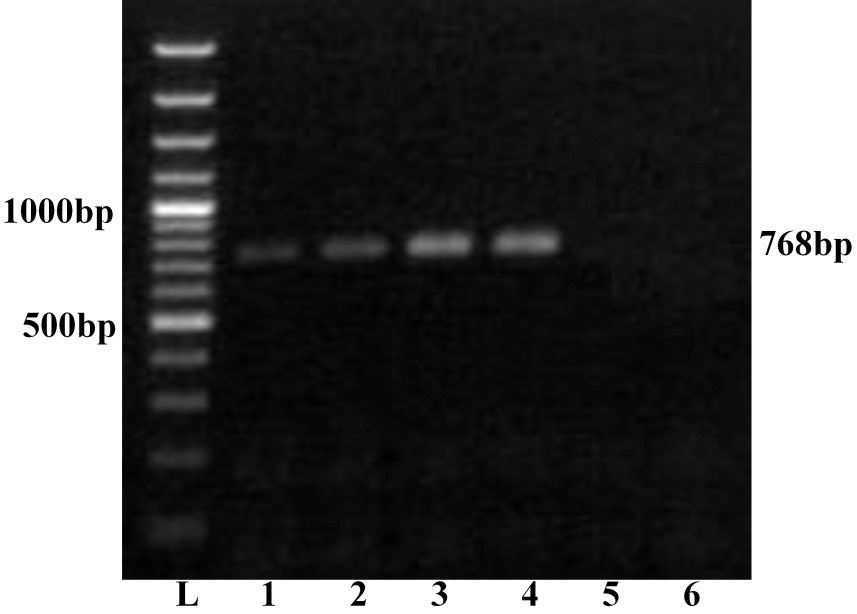
Further, all the samples were subjected to serotype specific RT-PCR using vp2 gene specific primers for all the BTV serotypes. The agarose gel electrophoresis showed a single expected 768bp amplification product with BTV16 vp2 gene primer only (Figure 2). However, remaining primers for other BTV serotypes did not show any amplification. Thus all the four samples were serotyped as BTV16. The PCR products of vp2 gene of all the four BTV samples were allowed for direct nucleic acid sequencing for confirmation of serotype. The BLASTN+ 2.2.31 search of nucleotide sequence of all the isolates showed maximum identity with vp2 gene of several isolates of BTV16 only. The vp2 gene serotype specific RT-PCR and nucleotide sequencing confirmed these isolates as BTV16. The nucleotide sequences of all the samples were submitted to GenBank database and accession numbers JN106018, JN106020, JN106021 and JN106022 were assigned for GNT27/IND, MBN48/IND, MBN50/IND and VJW66/IND isolates respectively.
Lane L: Ladder 100bp; 1: GNT27/IND; 2: MBN48/IND; 3: MBN50/IND; 4: VJW66/IND; 5: BHK21 cell control; 6: Nuclese free water control. The left side numbers indicate DNA marker and right side indicated size of PCR product.
Lane L: Ladder 100bp; 1: GNT27/IND; 2: MBN48/IND; 3: MBN50/IND; 4: VJW66/IND; 5: BHK21 cell control; 6: Nuclease free water control. The left side numbers indicate DNA marker and right side indicated size of PCR product.
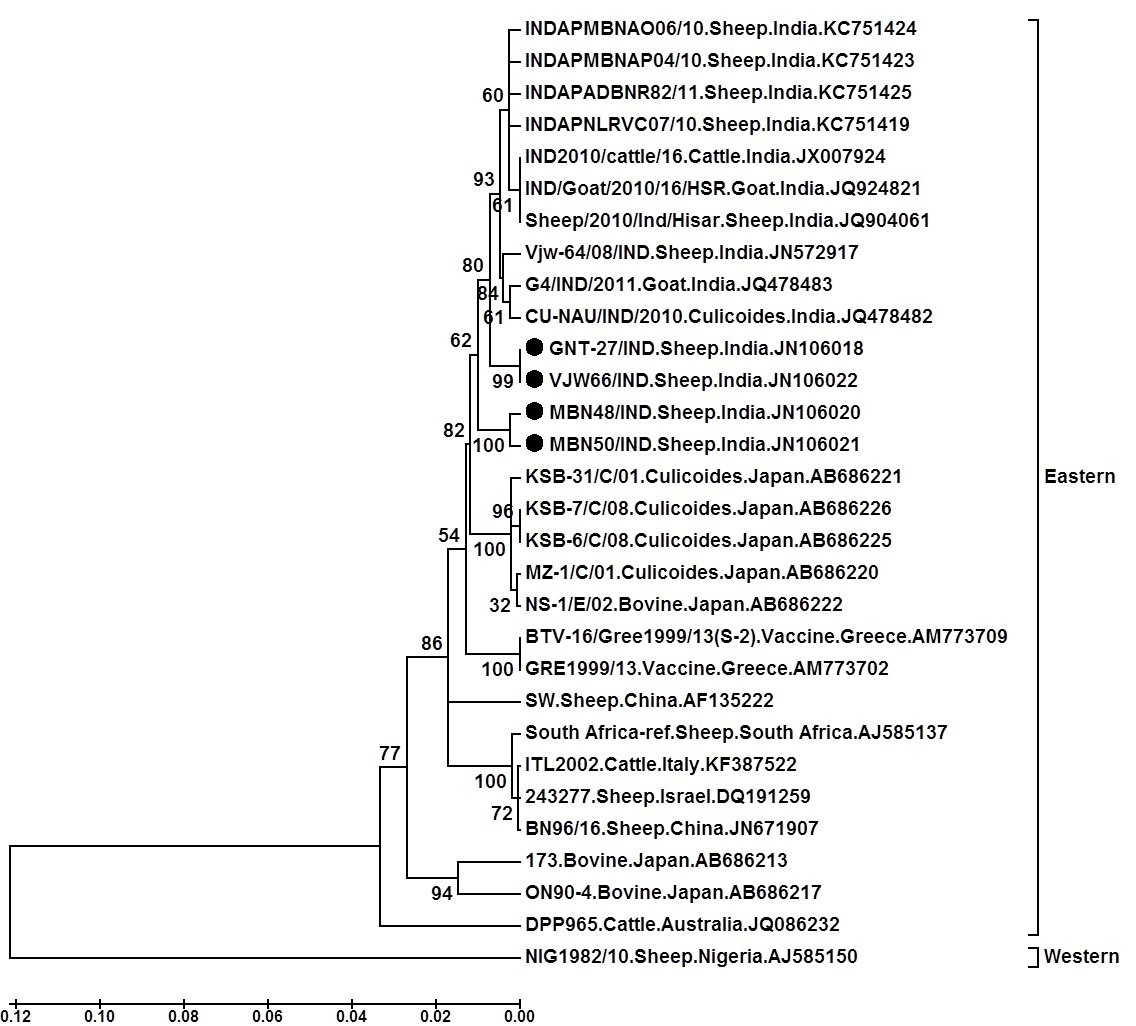
Tree was constructed using neighbour joining method with 1000 bootstrap values in Mega6 software programme (Tamura et al., 2013); ●=Selected isolate in this study
The sequence data of all the isolates were further analysed using Bioedit v7.2.3 (Hall, 1999) to calculate percent nucleotide as well deduced amino acid sequence identity with several other BTV16 isolates from India and different parts of the world (Figure 3). The sequence identity analysis revealed that isolates in study showed 92.6-97.4% nucleotide (nt) and 91.8-97.9% deduced amino acid (aa) identity among themselves, confirming that the isolates are different from each other. The sequence identity study of GNT27/IND, MBN48/IND, MBN50/IND and VJW66/IND (Accession numbers JN106018, JN106020, JN106021 and JN106022 respectively) with several other BTV 16 isolates (IND/Goat/2010/16/HSR, Vjw-64/08/IND, G4/IND/2011) (Minakshi et al., 2012; Shafiq et al., 2013; Dadawala et al., 2013) from India showed 99-91.6/99.5-91.3%, nt/aa identity. They also showed higher identity of 98.2-89.1/98.7-91.3%, nt/aa with BTV16 isolates from Japan (KSB-7/C/08, KSB-6/C/08, MZ-1/C/01, KSB-31/C/01, 173) (Shirafuji et al., 2012) and China (SW, BN96/16) (Yang et al., 2011). However, they showed slightly less identity (97.8-87.7/98.7-91.3%, nt/aa) with several other BTV16 isolates from Greece, Italy, Israel, South Africa and Australia.
The nucleotide sequence analysis from different geographical regions of India and abroad categorised BTV broadly in to ‘eastern’ or ‘western’ topotypes (Maan et al., 2010). The sequence analysis of GNT27/IND, MBN48/IND, MBN50/IND and VJW66/IND isolates (Accession numbers JN106018, JN106020, JN106021 and JN106022 respectively) showed overall 99-87.7/99.5-91.3%, nt/aa identity with eastern BTV16 viruses from India, Japan, China, South Africa, Italy and Israel. However, they showed only 71.0-75.9/81.1-87.6%, nt/aa identity with Western BTV16 (isolate NIG1982/10) from Nigeria (Mertens et al., 2013). Thus sequence identity study confirmed the eastern origin of isolates in study.
The phylogenetic analysis of nucleotide sequences using Mega 6 software programme formed two separate major, eastern and western clusters. The isolates in study formed a separate close cluster with other Indian BTV16 viruses in eastern cluster. They were also closely related to other eastern BTV16 isolates from Japan, Greece and china. Moreover, they were distantly related to eastern viruses from South Africa, Israel, Italy, Australia, China and Japan. However, western cluster having a single virus from Nigeria (Isolate NIG1982/10) (Mertens et al., 2013) was placed far apart from the isolates in study. The phylogenetic and sequence identity analysis revealed that isolates GNT27/IND (Accession number, JN106018), MBN48/IND (Accession number, JN106020), MBN50/IND (Accession number, JN106021) and VJW66/IND (Accession number, JN106022) are much closer to Indian, Japanese, Chinese or Greece BTV16 isolates.
Andhra Pradesh is one of the heavily BT infested state in India. Based on serum neutralization and virus isolation several different serotypes of BTV such as serotype 1, 2, 4, 6, 9, 10, 12, 13, 14, 16, 17, 18, 19 and 21 have been reported from Andhra Pradesh state (Prasad et al., 2009; Ranjan et al., 2013; Reddy et al., 2015). BTV16 was also isolated from sheep in Andhra Pradesh (isolate, Vjw-64/08/IND) and in goat from adjoining state of Tamil Nadu (isolate, IND/Goat/2010/16/HSR) (Minakshi et al., 2012; Shafiq et al., 2013). The isolates in study showed a high degree of identity (91.6-99/99.5-91.3%, nt/aa) with BTV16 from Andhra Pradesh (isolate, Vjw-64/08/IND) and Tamil Nadu (isolate, IND/Goat/2010/16/HSR). Since Andhra Pradesh and Tamil Nadu state are also endemic for Culicoides oxystoma, a known vector of BTV in India (Minakshi, 2010). Thus, BTV16 might be transmitted from one state to other state of South India either through migrating sheep population or vectors on animal body or through wind velocity. Several serotypes of BTV such as BTV1, 2, 10 and 16 were identified for BT outbreak in Andhra Pradesh in recent years. Due to serious outbreak of BTV16 in south India (Andhra Pradesh and Tamil Nadu) it is included in an inactivated Pentavalent vaccine formulation along with other serotypes 1, 2, 10 and 23 (Reddy et al., 2010). The knowledge about the prevalence of all the BTV serotypes and their genomic diversity is essential for the BTV control programme. Conventional serotyping along with modern molecular tests such as RT-PCR and nucleic acid sequencing can be used for surveillance of BTV serotypes prevalent in a particular geographical area. Based on serotype prevalence the appropriate vaccine can be selected.
Conclusion
BT is mainly a disease of sheep in India. There are 22 out of 26 serotypes which have been reported from the country. In this study four different BTV samples of sheep origin from Andhra Pradesh state were taken for vp2 gene based molecular characterization. All the samples were serotyped as BTV 16 based on vp2 gene serotype specific RT-PCR and its nucleic acid sequencing followed by GenBank data base search. The sequence identity and phylogenetic study showed that all the four BTV isolates are much closer to BTV16 isolates from Japan, China, Greece and other Indian BTV16 isolates. Therefore to control BT in the country suitable measures should be taken to control the import of animal and its products.
Competing interest
All authors declare that they have no conflict of interest.
Acknowledgements
Authors are thankful to ICAR, New Delhi for providing financial support under ‘All India network programme on Bluetongue’ and Department of Animal Biotechnology, Lala Lajpat Rai University of Veterinary and Animal Sciences, Hisar, Haryana for providing infrastructural facility.
Reference