Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (4): 199 – 203Real Time and Conventional PCR for Characterization of Salmonella sp. from Imported Meat to Egypt
Mohamed Sabry AbdElraheam Elsayed1, Eman Abdeeen1, Mohamed Ali Akiela2, Thaer Farouk3, Rasha Zahran1
- Department of Bacteriology, Mycology and Immunology faculty of veterinary medicine Sadat city University, Menoufyia governorate, Egypt
- Department of microbiology faculty of veterinary medicine Alexandria university
- Administration laboratories in Alexandria port-4, Egypt
*Corresponding author: [email protected]
ARTICLE CITATION:
Elsayed MSA, Abdeen E, Akiela MA, Farouk T, Zahran R (2014). Real time and conventional PCR for characterization of Salmonella sp. from imported meat to Egypt. Adv. Anim. Vet. Sci. 2 (4): 199 – 203.
Received: 2014–02–16, Revised: 2014–03–05, Accepted: 2014–03–09
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.4.199.203
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
This study was aimed to detect Salmonella spp. in imported frozen beef and buffalo meat cuts from India, Brazil, Australia and Newzeland. One hundred frozen meat samples were collected from supermarkets located at Alexandria province and from Alexandria port. Salmonella was biochemically identified in 18/100 (18%) samples. Serologically, S. paratyphi A and S. typhi were detected in 10/18 (55.56%) and 8/18 (44.44%) positive samples, respectively. Using conventional PCR, 18/18 (100%) were confirmed to be pathogenic by specific primer to rfbS gene of the pathogenic strains of Salmonella responsible for the most food poisoning while all of these pathogenic isolates 18/18 (100 %) were also confirmed by real time PCR amplifying the invA gene of Salmonella spp.
INTRODUCTION
Microbial contamination of food is serious problem and trading between countries raises the liability for outbreaks. Food borne illnesses are considered by the World Health Organization (WHO) as diseases either infectious or toxic made by causative agents in ingested food. The reports in 2005 recorded 1.8 million people died from diseases causing diarrhea and high proportion of which was attributed to contamination of food and drinking water (WHO, 2007). Recently, some food borne diseases are considered as emerging diseases. Various food borne pathogens have been identified for food borne illness. Campylobacter, Escherichia coli O157:H7, Listeria monocytogenes and Salmonella are found to be responsible for most of food origin outbreaks (Chemburu et al., 2005). Mainly, many foods are source of food borne outbreaks if these are contaminated with Salmonella spp. These foods are: undercooked eggs, poultry, meat, raw milk, dairy products, seafood, chocolate, salad and spices. The reported infective dose of about 15–20 microorganisms are sufficient to cause pain in stomach. Mostly diarrhea, nausea, chills, fever, and headache appears after incubation period of 12 to 24 hrs (Abadias et al., 2008). Sometimes fatal infections of adults and children can occur from typhoid and paratyphoid fever due to bacteremia and destructive inflammation of the intestine and other organs. Traditional and conventional methods for identification of microbial pathogens rely mainly upon specific bacteriological and biochemical identification. Culture and isolation have high reliability for accurate detection of foodborne pathogen. Although culture based methods are considered to be standard techniques for detection of single bacteria like S. aureus, Salmonella, Coliforms , E.coli, etc. (Ayçiçek et al., 2004), however they are labour intensive and time consuming as it take 3 days to obtain the results. Implementation of immunological methods for detection of bacterial agents, spores, toxins and viruses expressed great success (Iqbal et al., 2000). Monoclonal antibodies have been used in the diagnosis of food borne pathogens as Salmonella enterica (Schneid et al., 2005), Salmonella typhi (Kumar et al., 2003). Most of immunological tests used for detection of foodborne pathogens are agglutination test (Matar et al., 1997) western blot test (Rasooly and Rasooly, 1998), enzyme immunoassay (EIA) (Borck et al., 2002), enzyme linked immunosorbent assay (ELISA) (Bennett, 2005). The sensitivity and specificity of the polymerase chain reaction (PCR) is very high as it is able to detect even single bacterium that's because it detects a single copy of targeted sequence of DNA, amplification of the target rather than the signal makes it promising, and lowers the liability of false–positive results as well as lower time required for performing the technique (Batt, 2007). Identification and differentiation between Typhi and Paratyphi A using rfbS primers targeting the gene prt that encodes CDP–paratose synthase, which essentially converts CDP–4–keto–3,6–dideoxyglucose to CDP–paratose. The gene prt is present in both serovars (Hirose et al., 2002). Salmonella virulence mainly relies on chromosomal and plasmid factors. Chromosomally situated invasion gene invA is thought to enable the invasion of salmonellae into cultured epithelial cells (Galan and Curtiss, 1989). The invA gene is present on Salmonella pathogenicity island 1 (SPI–1) that enables salmonella in invading epithelial cells. This gene is highly conserved in almost all Salmonella serotypes and has been used as a potential target for Salmonella detection (Li et al., 2012).
MATERIALS AND METHODS
Preparation of Samples (APHA, 1982)
25 grams of frozen meat sample was transferred to 225 ml of sterile peptone water (should be representative and taken from the different parts of the carcass), with application of hygienic measures.
Isolation of Salmonella Spp
- a) Pre–enrichment onto un–selective liquid buffered peptone water, incubation at 37ºC for 24hrs.
- Enrichment onto Rappaport–Vasiliadis soya broth is incubated at 41.5ºC for 24hrs and Muller–kauffmann tetrathionate/novobiocin broth alternatively 37ºC for 24 hrs.
- Plating out of Salmonella from the cultures obtained in the previous step, onto Xylose lysine Deoxycholate (XLD) agar and Hektoen enteric agar (HK) then incubated at 37ºC for 24hours,
- Purification: all typical or suspect colonies streaked onto the surface of nutrient agar sloops then incubate at 37ºC for 24hours.
Identification of Salmonella Isolates
- a) Biochemical identification using Triple Sugar Iron agar, Urea agar, L–lysine Decarboxylation medium according to (koneman et al., 1992).
- Serological Identification of Salmonella isolates according to the method of Edwards and Ewing, 1972.
Molecular Identification of Salmonella Isolates
Extraction of genomic DNA was done according to the method of Sambrook et al., 1989:
- Preparation of the overnight culture: One colony from each strain grown on each media was picked up and cultured in 10 ml Lauria bertani broth and incubated at 37ºC overnight in shaker incubator.
- Preparations of the bacterial isolates for DNA extraction: overnight cultured cells cooled on ice for 10 minutes, centrifuged at 4°C for 5 minutes to be pelleted down, resuspended in 0.5 ml Tris EDTA buffer, allowed for 2 cycles freezing and thawing. The cells were then incubated 1 hour at 37oC with 1μl lysozymes (final concentration 100μg/ml). Proteinase–K was added 1μl/0.5 ml (final concentration 100μg/ml) and incubated for further 1 hour at 56oC with shaking.
- Trizol extraction: one milliliter trizol was added and after 5 minutes of incubation at room temperature, chloroform 0.4 ml added, vortexed for 15 seconds, and kept for 3 minutes at room temperature. Then it was centrifuged at 14000 rpm for10 minutes at 4oC. The upper liquid layer containing RNA was completely removed; the DNA in the interphase was precipitated with 0.6 ml absolute ethanol and kept for 3 minutes at room temperature before centrifugation at 4000 rpm for 5 minutes at 4oC. The supernatant removed and the pelleted DNA washed twice with sodium citrate 0.1 in 10% ethanol. The DNA pellet was kept in the washing solution at room temperature for 30 minutes with periodical mixing, and then centrifugation was done at 4000 rpm for 5 minutes at 4oC. Following the 2 washes, the DNA was resuspended in 2ml of 75% ethanol, kept at room temperature for 20 minutes with periodical mixing and then centrifuged. The DNA pellet was finally dried briefly for 5 minutes under vacuum and dissolved in 50μl of 8 mM NaOH. The pH was then adjusted at 8 by adding 115 µl/ml of 0.1 M HEPES. Two µl of RNAase were then added and incubated at 37oC for 1 hour.
- Purification of DNA: DNA was mixed with 1ml of wizard DNA clean up resin in 1.5 ml microfuge tube. Resin/DNA mix was then transferred to the minicolumn provided with the kit and the solution was drained out by adding slight pressure over the minicolumn. Resin/DNA mix was then washed with 2ml of 80% isopropanol and dried by centrifugation at 14,000 rpm/ 2 minutes at 4oC. DNA was then eluted with 50μl of hot 80oC TrisEDTA buffer.
Polymerase Chain Reaction
The primers used were as follows: forward (5'–CTT GCT ATG GAA GAC ATA ACG AAC C–3') reverse (5'–CGT CTC CAT CAA AAG CTC CAT AGA–3'). Primers GenBank accession no. M29682 synthesized by BioBasic, Canada, purified by HPLC and concentrated to 100 pg/μl. Each reaction volume was 50µl.The mixture contained 0.3µM of each primer, 200 µM of dATP, dCTP, and dGTP, 190µM dTTP, 10µM DIG–11–dUTP (Roche Diagnostics), 0.5 U of Taq polymerase, 5µl of 10xPCR buffer, and 1.5 mM of MgCl2 with various concentrations (from 50ng to 0.5pg) of genomic DNA. The PCR program conducted was as following: 94ºC for 5 min, followed by 5, 8 or 30 cycles at 94, 60 and 72ºC respectively, each for 1 min and the final extension at 72ºC for 5 minutes. Then, 5µl of every product loaded into wells of 2% agarose gel with 0.5µg/ml ethidium bromide. Molecular weight marker 100bp ladder (Fermentas) was used. PCR products were electrophoresed, visualized with UV light and images stored using a gel documentation system (Gel Doc1000; BioRad, Hercules, CA) expect product size (bp) 258 bp (Hirose et al., 2002).
Real time qRT–PCR utilizing primers and probe targeting invA gene were as follows: forward primer 5’–GCG TTC TGA ACC TTT GGT AAT AA–3’ reverse primer 5’–CGT TCG GGC AAT TCG TTA–3’ Sal–TM (probe) 5’–FAM–TGTTG CGGTG GGTTT GTTGTCTT– TAMRA–3’ Synthesized by BioBasic, Canada, purified by HPLC and concentrated to 100pg/ul. The primers (GenBank accession no. NC_006511.1) and probe (GenBank accession no. NC 015761.1) amplify the invA gene of Salmonella spp. The quantitation was done by Q–PCR using probe based method. The PCR mix was performed as follow : 2X reagent Mix 2.5 µl, 1.0 µl of magnesium chloride sol 25mM, 0.01 µl forward primer, 0.01 µl reverse primer, Probe 0.01µl, DNA 0.5 µl, RNAase– DNase free water To 25 µl. The reaction was done using agilant MX3005 probe real time PCR. The program was adjusted as follow: thermo start activation step 95°C for 10 min 1 cycle, and 30 cycles consisted of denaturation 95°C for15 sec, annealing 57°C for 30 sec and extension 60°C for 30 sec .The immersion light created by the hydrolysis of the Taq man probe was read after the end of each extension step. Interpretation of results was made with the help of the real–time PCR system user guide instructions. Briefly, in every PCR assay Ct value >16.9 for the invA probe considered positive and zero result of Ct values (non–amplified, NA) considered negative, avoidance of PCR inhibitors made by repeating PCR assay with tenfold diluted DNA template suspension (Novinscak et al., 2007).
Data Analysis The prevalence to every test calculated after dividing number of positive samples by number of all samples under test within the specified period.
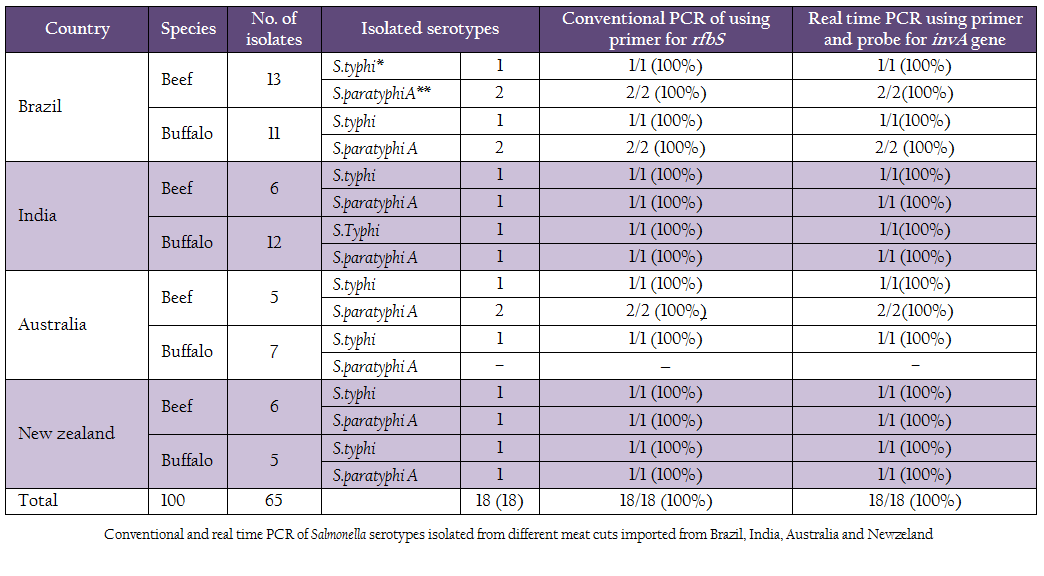
Table 1: Conventional and real time PCR of Salmonella serotypes isolated from different meat cuts imported from Brazil, India, Australia and Newzeland.
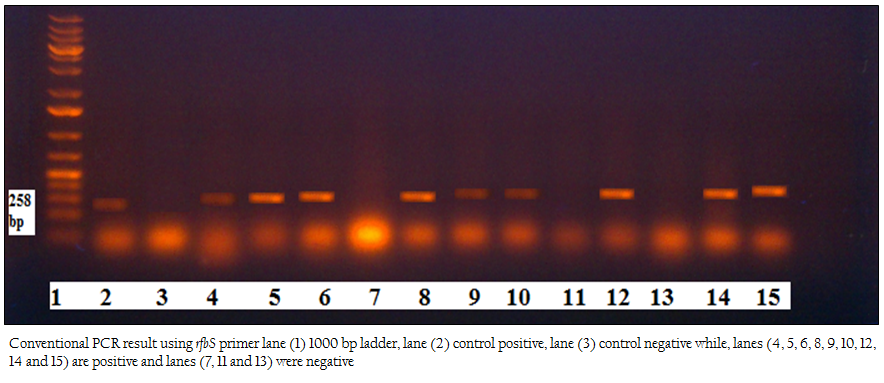
Figure 1: Conventional PCR result using rfbS primer lane (1) 1000 bp ladder, lane (2) control positive, lane (3) control negative while, lanes (4, 5, 6, 8, 9, 10, 12, 14 and 15) are positive and lanes (7, 11 and 13) were negative
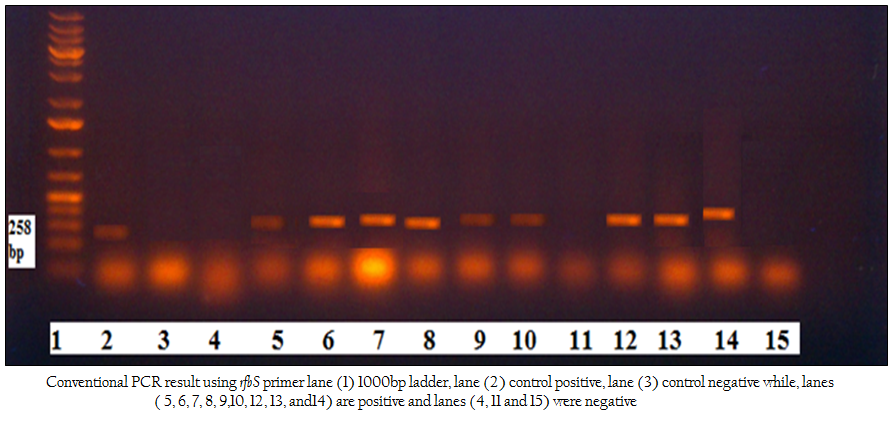
Figure 2: Conventional PCR result using rfbS primer lane (1) 1000bp ladder, lane (2) control positive, lane (3) control negative while, lanes ( 5, 6, 7, 8, 9,10, 12, 13, and14) are positive and lanes (4, 11 and 15) were negative
RESULTS
One hundred frozen meat samples were collected from supermarkets located at Alexandria province and from Alexandria port. Salmonella was biochemically identified in 18/100 (18%) samples. Serologically S. paratyphi A and S. typhi were detected in 10/18 (55.56%) and 8/18 (44.44%) of the positive samples, respectively. Using Conventional PCR, 18/18 (100%) were confirmed to be pathogenic by specific primer to rfbS gene of the pathogenic strains of Salmonella responsible for the most food poisoning while, all of these pathogenic isolates 18/18 (100 %) were confirmed by real time PCR using primers and probe that amplify the invA gene of Salmonella spp Table 1; Figure 1, Figure2).
DISCUSSION
Historically meat, poultry and eggs are considered as major sources of high quality animal protein. Potentially they may harbor, or become environmentally contaminated with certain pathogenic microorganisms during preharvest production or processing throughout the food chain (Forsythe, 1996).Therefore; these food types have been considered vehicles for salmonellosis transmission to human beings. (Dallal et al., 2009).
Therefore, our study focused on isolation and identification of Salmonella spp. from meat cuts imported from (Brazil, India, Australia and Newzeland) to Egypt. Salmonellosis is serious zoonotic disease of great public health concern because of its endemic nature, higher morbidity levels and being associated with a wide range of foods. Results obtained in Table 1 which mainly represent the prevalence of isolated and biochemically identified Salmonella was 18/100 (18%) (Table 1) found in our study are in agreement with the result found by (D’Aoust et al., 2000), who reported that salmonella prevalence in post–slaughter beef ranges from 0.6 to 20.3% in about 23045 samples tested from Denmark, Germany, Nigeria, Portugal and USA. Also prevalence of Salmonellae in meats at retail level of raw beef ranges from 1.3 to 21.5% in about 3743 samples tested in Denmark, Japan, Mexico, Thailand and Netherlands. Also, similar prevalence of Salmonella spp. was reported by Dallal et al., 2010 from beef in Iran with prevalence of 38/189 (20.1%) and (Yang et al., 2010) who reported prevalence of Salmonella from beef in China was 13/78 (16.7%).
The studies on the serotyping identified S.typhi* with percentage of 8/18 (44.44%) and S.paratyphi A** with percentage of 10/18 (55.56%) These data nearly comply with that obtained by Jegadeeshkumar et al., 2010 who reported Salmonella prevalence from meat was 20% and the serotype isolated after examining three food types (fish, fruits and meat) were S.typhi, S.paratyphi A ,S.paratyphi B and S.typhimurium 55.5%, 48.1%, 25.9% and 22.2% respectively in positive samples.
Salmonella typhi has well adapted and is responsible for causing deadly invasive typhoid fever in human beings leading to high morbidity and mortality. Salmonella Paratyphi A dT+ since the end of the 1990s has become increasingly prominent. Many outbreaks associated with this variant in France (Desenclos et al., 1996) in Canada (Gaulin et al., 2002) in Australia and several European countries (Denny et al., 2007) have been recorded. High prevalence of isolation of the multi–drug resistant salmonella paratyphi A dT+clone has been recorded from poultry and poultry products in Germany and Netherlands (Miko et al., 2002). Contamination of meat with bacterial pathogens has been reported in many countries (Kinsella et al., 2008). Others recognized open air bacterial spoilage of meat by presence of gram–negative organisms (Eribo and Jay, 1985). Unhygienic abattoirs environment and practices during post–process handling has serious impact on health (Abdullahi et al., 2006). Not only environment, but also the animals will be slaughtered can constitute contamination source (Sofos et al., 2000). Live animals harboring the pathogens and contaminated environment constitute sources for contamination during slaughtering and meat products during processing, storage and handling. In addition, role of animals in contaminating the carcass was confirmed by (Arthure et al., 2008) who found that lymph nodes of culled cattle meat contain high prevalence of Salmonella than did those from fed cattle carcasses consequently liability of increasing number of Salmonella in carcass is present. Additionally, butcher hands, dress and slaughtering equipments have also been considered as other contamination sources (Aftab et al., 2012). Serotyped isolates were confirmed to be Salmonella with conventional PCR using rfbS Primer and real time PCR using primer and probe for invA gene with percentage of 18 /18(100%). The sensitivity of PCR in detection of Salmonella was confirmed by Mccarthy et al., 2009 who optimized multiplex PCR assay and quantitative real–time PCR assay for detection and differentiation of S. enterica Typhimurium and Heidelberg in foods. The multiplex PCR assay detected S. Enterica isolates at concentrations as low as 1 CFU/g of inoculated Cheddar cheese, raw turkey, and cooked turkey. They concluded that PCR has significantly saved time needed to identify S.enterica Typhimurium and Heidelberg, making this rapid, sensitive, specific and selective diagnostic tool. Also higher PCR sensitivity was confirmed by Robles et al., 2009 who detected 4 Salmonella containing meat samples out of 50 using PCR which were more than culture technique that detected only 3 out of 50, therby confirming higher sensitivity than cultivation.
CONCLUSION
Periodical testing of imported meat is of great importance so as to prevent foodborne outbreaks. Furthermore traditional methods should be substituted by advanced molecular methods for detection of serious bacterial agents because they are fast, sensitive and labor saving.
AUTHOR CONTRIBUTIONS
Dr Mohamed Elsayed shared in PCR identification, data analysis and writing this paper, Dr Eman Abdeen shared in isolation, Dr Akiela and Dr Rasha Zahran shared in paper revision while Dr Thaer Farouk helped in finishing PCR.
ACKNOWLEDGEMENTS
We thank the animal health research institute Dokki Giza Egypt unit of molecular biology for their help in performing PCR.
REFERENCES
Abadias M, Usall J, Anguera M, Solson C, Vinas I (2008). Microbiological quality of fresh, minimally–processed fruit and vegetables, and sprouts from retail establishments. Intl. J. Food Microbiol. 123: 121 – 129.
http://dx.doi.org/10.1016/j.ijfoodmicro.2007.12.013
PMid:18237811
Abdullahi IO, Umoh VJ, Ameh JB, Galadima M (2006). Some hazards associated with the production of a popular roasted meat (tsire) in Zaria, Nigeria. Food Control. 17(5): 348 – 352.
http://dx.doi.org/10.1016/j.foodcont.2004.11.010
Aftab M, Rahman A, Qureshi MSS, Akhter U, Sadique A, Zaman S (2012). Level of Salmonella in beef of slaughtered cattle at Peshawar. J. Anim. Plant Sci. 22 (2): 24.
American public health association (1982). Compendium of methods for microbiological examination of foods. 1015 fifteen street, N.W, washington, D.C., pp: 23 – 35.
Arthure TM, Harhay BDM, Bosilevac JM, Guerini MN, Kalchayanand N,Wells JE, Shackelford SD, Wheeler TL, Koomaraie M (2008). Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J. Food Prot. 71(8): 1685 – 1688.
Ayçiçek H, Aydogan H, Küçükkaraaslan A, Baysallar M, Basustaoglu AC (2004). Assessment of the bacterial contamination on hands of hospital food handlers. J. Food Cont. 15: 253 – 259.
http://dx.doi.org/10.1016/S0956-7135(03)00064-1
Batt CA (2007). Food pathogen detection. Sci. 316: 1579 – 1580.
http://dx.doi.org/10.1126/science.1140729
PMid:17569853
Bennett RW (2005). Staphylococcal enterotoxin and its rapid identification in foods by enzyme–linked immunosorbent assay based methodology. J. Food Prot. 68: 1264 – 1270.
PMid:15954720
Borck B, Stryhn H, Ersboll AK, Pedersen K (2002). Thermophilic Campylobacter spp. in turkey samples: evaluation of two automated enzyme immunoassays and conventional microbiological techniques. J. App. Microbiol. 92: 574 – 582.
http://dx.doi.org/10.1046/j.1365-2672.2002.01568.x
PMid:11872135
Chemburu S, Wilkins E, Abdel–Hamid I (2005). Detection of pathogenic bacteria in food samples using highly–dispersed carbon particles. Biosens Bioelectron. 21: 491 – 499.
http://dx.doi.org/10.1016/j.bios.2004.11.025
Dallal MMS, Taremi M, Gachkar L, Modarressi S, Sanaei M, Bakhtiari R, Yazdi M KS, Zali MR (2009). Characterization of antibiotic resistant patterns of Salmonella serotypes isolated from beef and chicken samples in Tehran Jundishapur. J. Microbiol. 2(4): 124 – 131.
Dallal SMM, Doyle MP, Rezadehbashi M, Dabiri H, Sanaei M, Modarresi S, Bakhtiari R, Shariiy K, Taremi M, Zali MR, Sharii–Yazdi MK (2010). Prevalence and antimicrobial resistance profiles of Salmonella serotypes, Campylobacter and Yersinia spp. Isolated from retail chicken and beef Tehran Iran. J. Food Cont. 21: 388 – 392.
http://dx.doi.org/10.1016/j.foodcont.2009.06.001
D'Aoust JY (2000). Salmonella; Chapter 45 in "The microbiological safety and quality of food."(Lund B M, Baired–Parker A C and Gould GW, eds.), Vol ll, pp: 1233 – 1299.
Denny J, Threlfall J, Takkinen J, Lofdahl S, Westrell T, Varela C, Adak B, Boxall N, Ethelberg S, Torpdahl M, Straetemans M, vanPelt W (2007). Multinational Salmonella Paratyphi B variant Java (Salmonella Java) outbreak. European Surveillance. 12: 2 – 6.
PMid:18179762
Desenclos JC, Bouvet P, Benz–Lemoine E, Grimont F, Desqueyroux H, ebiere I, Grimont PA (1996). Large outbreak of Salmonella enterica serotype Paratyphi B infection caused by a goats' milk cheese, France, 1993: a case finding and epidemiological study. J. British Microbiol. 312: 91 – 94.
Edwards PR, Ewing WH (1972). Identification of Enterobacteriacea 3rd edn. Burgess Publishing Co. Minneopolis. USA.pp: 241 – 261.
Eribo BE, Jay JM (1985). Incidence of Acinetobacter spp. and other Gram–negative bacteria in fresh and spoiled ground beef. Appl. Envir. Microbiol.49: 256 – 257.
PMid:3977314 PMCid:PMC238385
Forsythe RH (1996). Food safety: a global perspective. Poult. Sci. 75(12): 1448 – 1454.
http://dx.doi.org/10.3382/ps.0751448
PMid:9000265
Galan JE, Curtiss R (1989). Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Nat. Acad. Sci. USA. 86: 6383 – 6387.
http://dx.doi.org/10.1073/pnas.86.16.6383
PMid:2548211 PMCid:PMC297844
Gaulin C, Vincent C, Alain L, Ismail J (2002). Outbreak of Salmonella paratyphi B linked to aquariums in the province of Quebec, Canada. Commun. Dis. Rep. 28: 89 –91.
Hirose K, Itoh K, Nakajima H, Kurazono T, Yamaguchi M, Moriya K, Ezaki T, Kawamura Y, Tamura K, Watanabe H (2002). Selective Amplification of tyv (rfbE), prt (rfbS), viaB, and fliC Genes by Multiplex PCR for Identification of Salmonella enterica serovars typhi and paratyphi A. J. Clin. Microbiol. 40(2): 633 – 636.
http://dx.doi.org/10.1128/JCM.40.02.633-636.2002
PMid:11825983 PMCid:PMC153373
Iqbal SS, Mayo MW, Bruno JG, Bronk BV, Batt CA, Chambers JP (2000). A review of molecular recognition technologies for detection of biological threat agents. Biosens Bioelectron. 15: 549 – 578.
http://dx.doi.org/10.1016/S0956-5663(00)00108-1
Kumar JD, Saritha V, Moorthy K, Kumar SBT (2010). Prevalence, antibiotic resistance and RAPD analysis of food isolates of Salmonella species. Intl. J. Biological Technol. 1(3): 50 – 55.
Kinsella KJ, Prendergast DM, McCann MS, Blair IS, McDowell DA, Sheridan JJ (2008). The survival of Salmonella enteric serovars typhimurium DT 104 and total viable counts on beef surfaces at different relative humidities and temperatures. J. Appl. Microbiol. 106: 171 – 180.
http://dx.doi.org/10.1111/j.1365-2672.2008.03989.x
PMid:19054240
Koneman EW, Allen SD, Janda WH, Schrechenberger PC, Winn WC (1992). Color atlas and text book of diagnostic microbiology (fourth edition), 1979, 1983, 1988, 1992 by Lippincott Company.
Kumar S, Balakrishna K, Tuteja U, Batra HV (2003). Application of monoclonal antibodies to flagellin of Salmonella typhi for its rapid detection in foods. Indian J. Microbiol. 43: 193 – 197.
Li Q, Cheng W, Zhang D, Yu T, Yin Y, Ju H, Ding S (2012). Rapid and sensitive strategy for Salmonella detection using an InvA gene–based electrochemical DNA sensor. Intl. J. Electrochemical Sci. 7: 844 – 856.
Matar GM, Hayes P S, Bibb WF, Bala S (1997). Listeriolysin O–based latex agglutination test for the rapid detection of Listeria monocytogenes in foods. J. Food Prot. 60: 1038 – 1040.
Mccarthy N, Reen J F, Buckley Fj, Frye J, Jonathan G E, Boyd F, Gilroy D (2009). Sensitive and rapid molecular detection assays for Salmonella enterica serovars typhimurium and heidelberg. J. Food Prot. 72(11): 2350 – 2357.
PMid:19903399
Miko A, Schroeter BA, Dorn C, Helmuth R (2002). Molecular characterization of multi resistant d–tartrate positive Salmonella enterica serovar paratyphi A isolates. J. Clin. Microbiol. 40: 3184 – 3191.
http://dx.doi.org/10.1128/JCM.40.9.3184-3191.2002
PMid:12202551 PMCid:PMC130825
Novinscak A, Surette C, Filion M (2007). Quantification of Salmonella spp. in composted biosolids using a TaqMan qPCR assay. J. Microbiol. Meth. 70: 119 – 126.
http://dx.doi.org/10.1016/j.mimet.2007.03.019
PMid:17481755
Rasooly A, Rasooly RS (1998). Detection and analysis of Staphylococcal enterotoxin A in food by western immunoblotting. Intl. J. Food Microbiol. 41: 205 – 212.
http://dx.doi.org/10.1016/S0168-1605(98)00050-6
Robles MAG, Loredo MA, Lvarez–Ojeda GA, Osuna–Garcia J A, Matr´ inez IO, Morales–Ramos LH, Fratamico P (2009). PCR detection and microbiological isolation of Salmonella spp. from fresh beef and cantaloupes. J. Food Sci. 74(1): 37 – 40.
http://dx.doi.org/10.1111/j.1750-3841.2008.01006.x
PMid:19200105
Schneid AD, Ludtke CB, Diel C, Aleixo JAG (2005). Production and characterization of monoclonal antibodies for the detection of Salmonella enterica in chicken meat. Brazilian J. Microbiol. 36: 163 –169.
http://dx.doi.org/10.1590/S1517-83822005000200012
Sofos JN, Pearson AM, Dutson TR (2000). Microbial growth and its control in meat, poultry and fish, Blackie academic and professional, Glasgow, UK. pp: 359 – 403.
World Health Organization (2007). Food safety and food–borne illness. fact sheet no. 237,Geneva.
Yang B, Qu D, Zhang X, Shen J, Cui S, Shi Y, Xi M, Sheng M, Zhi S, Meng J (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Intl. J. Food Microbiol. 141(2): 63 – 72.
http://dx.doi.org/10.1016/j.ijfoodmicro.2010.04.015
PMid:20493570





