Advances in Animal and Veterinary Sciences
Research Article
Molecular Identification of Babesiosis in Stray Dog at Chittagong Metropolitan Area, Bangladesh
Md. Nur-e-Azam1*, Md. Shafiqul Islam1, Motahara Tasneem2, Tofazzal Md. Rakib1, Md. Ekramul Bari1, Md. Abdul Mannan3, Mohammad Mahbubur Rahman, Md. Masuduzzaman, Mohammad Alamgir Hossain1
1Departmentt of Pathology and Parasitology; 2Department of Microbiology and Veterinary Public Health; 3Department of Medicine and Surgery, Chittagong Veterinary and Animal Sciences University, Bangladesh.
Abstract | An epidemiological study of babesiosis in stray dogs was conducted at Chittagong Metropolitan area, Bangladesh, for six months. Blood samples from 130 stray dogs were collected from nine randomly selected areas along with the information of age and sex. Blood samples were initially examined by Giemsa’s stained blood smear method and DNA was extracted and Babesia spp. were confirmed by amplifying 18S rRNA gene. The prevalence was determined 6.92% in microscopic techniques and 4.61% by PCR. Significantly higher frequency of babesiosis was found in adult (10.11%) and male dog (11.94%) than younger and female, respectively (p<0.05). Furthermore, we sought to develop a seminested PCR to detect and differentiate Babesia gibsoni (Asia genotype) from other Babesia sp. An outer primer pair was designed to amplify a ~340 bp fragment of the 18S rRNA genes. Forward primers were premeditated that would particularly amplify a smaller fragment from each organism in the seminested PCR. Seminested PCR revealed that, all parasitic isolates were identified as Babesia gibsoni (Asian type). Sequencing of the PCR products and analysis of sequence homology with other Babesia using NCBI BLAST our isolate CVASU/BD/Babesia (KR349267) showed 95% nucleotide homology with other Bangladeshi Babesia gibsoni isolate BD 37 (LC008285) and Kolkata isolate (KJ142323). The phylogenetic tree shows our isolate is most closely related to but distinct from Kolkata isolate. CVASU/BD /Babesia and Kolkata isolate forms clade which indicates geographical location may have some influence on the distribution of the parasite. These results recommended that Babesia gibsoni (Asian type) parasites are widespread in stray dog in Chittagong Metropolitan area, Bangladesh. Further investigations are required to clarify the origin, vector and pathogenesis of these parasites tracking in dogs in Bangladesh.
Keywords | Stray dog, 18S rRNA gene, Babesia gibsoni, Seminested PCR, Sequencing, Phylogenetic tree
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | October 26, 2016; Accepted | November 07, 2016; Published | November 22, 2016
*Correspondence | Md. Nur-e-Azam, Departmentt of Pathology and Parasitology, Chittagong Veterinary and Animal Sciences University, Bangladesh; Email: nnureazam@yahoo.com
Citation | Nur-e-Azam M, Islam MS, Tasneem M, Rakib TM, Bari ME, Mannan MA, Rahman MM, Masuduzzaman M, Hossain MA (2016). Molecular identification of babesiosis in stray dog at Chittagong metropolitan area, Bangladesh. Adv. Anim. Vet. Sci. 4(11): 604-612.
DOI | http://dx.doi.org/10.14737/journal.aavs/2016/4.11.604.612
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Nur-e-Azam et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bangladesh has a wide range of climatic region, from plane and coastal to the hilly area, which make it suitable for a varied range of vectors and pathogens of medical and veterinary meaning, whose transmission and geographical sharing are closely allied to regional temperature, rainfall and humidity (Rahman, 2014; Nath et al., 2016). The vector borne parasitic diseases affect not only human but also the domestic, wild and companion animals (Rani et al., 2010). Stray dogs are unconfined dogs that live almost wherever cities exist and the local human inhabitants permit (Daniels, 1983; Das et al., 2011). They act as the usual connectors between people and nature (Beck, 1973) and associated with >60 zoonotic diseases among which can pose serious health concern, as well as significant economic impact from veterinary perspective (Ranjbar-Bahadori et al., 2008; Das et al., 2012; Rakib et al., 2016).
Parasitism is the most frequently encountered diseases in dogs all over the world (Traub, 2003; Azam et al., 2015). Developing countries like Bangladesh, the number of stray dogs that coexist with human being is high in most cities and villages which contain a potential risk of infections for human beings. The distribution and concentration of parasitism in dogs are predisposed by geological, climatic, cultural and economic factors (Robertson et al., 2000; Das et al., 2012). Amongst the diverse ubiquitous canine vector-borne diseases, canine babesiosis is very common and clinically significant disease caused by protozoa which are intra-erythrocytic and belong to genus Babesia (Singh et al., 2014; Azam et al., 2015). Canine babesiosis, a tick-borne infectious disease of dogs, has been illustrated globally as an emerging veterinary problem (Duh et al., 2004; Irwin, 2009). The escalating number of canine Babesia species, geographical allocation, changeable tick vectors and modes of transmission often consequence in varied of pathogenic and clinical presentations (Ayoob et al., 2010).
Regarding the diagnosis of canine babesiosis, direct microscopic assessment of the stained blood smear is the most frequently used technique as it is convincing, realistic, and cost effective diagnostic method but not essentially detects parasites in dogs with barely visible or chronic infections since the level of parasitemia is near to the ground (Cacciò et al., 2002). The serological methods, enzyme linked immunosorbent assay (ELISA) and indirect fluorescent antibody test (IFAT) for B. gibsoni parasites, stand very sensitive but reasonably specific because of antigenic cross-reactions to B. canis (Yamane et al., 1993) and normal dog erythrocytes (Yamane et al., 1993; Adachi et al., 1994). Therefore, the advancement of sensitive as well as highly specific system for the diagnosis of canine babesiosis still cares concern. In this regard, recent progress in molecular biology techniques resembling polymerase chain reaction (PCR) have made it possible to detect and identify piroplasms with better specificity and sensitivity than traditional methods (Birkenheuer et al., 2003b; Jefferies et al., 2009; Salem and Farag, 2014; Singh et al., 2014).
Regarding Bangladesh scenario, although there are periodic reports of canine babesiosis based on conventional diagnostic methods (Tarafder and Samad, 2012; Mahmud et al., 2014), the true condition of canine babesiosis is still not understandable barring only two reports (Talukder et al., 2013; Terao et al., 2015) employing the PCR based assays. At present, there are no statistics available on the distribution, prevalence, parasitic burden and risk factors associated with babesiosis of stray dogs in Chittagong. Furthermore, molecular detection of canine babesiosis has not yet been explored from Chittagong of Bangladesh, so the present work was carried out 1: To know the prevalence of canine babesiosis and association with risk factors (Age, Sex, and Location) in this part of the country through PCR based assays 2: Capacity development for genotyping/subgenotyping of Babesia spp. at Chittagong Veterinary and Animal Sciences University (CVASU).
Materials and methods
The study was conducted in the Chittagong Metropolitan area (CMA) of Bangladesh. Among the total CMA, nine (9) areas were randomly selected for the sampling. They are (Panchlaish, Chawkbazar, Muradpur, Alankar, Kattoli, Pahartali, Bayzid, Ambagan and New market). The area were chosen on the basis of high density of stray dog in CMA (Das et al., 2012). Moreover, selection of study areas were based on probability of high prevalence of different vectors, the climatic condition and geographical location which might favours the occurrence of such diseases. The study was undertaken for a period of 6 months from July 2014 to December 2014. The survey was conducted as cross sectional study. Stray dogs from the Chittagong Metropolitan area were selected for this study as target sample. To determine the age susceptibility to babesiosis the stray dogs were categorized as young (below one year) and adult (above one year). The approximate age of the stray dogs was estimated by examining the teeth. Dogs having all white and shiny permanent teeth without worn off cups on the incisors were considered as young (below one year) and dogs having teeth yellowish discoloration and tarter formation with worn cups on the incisors were considered as (above one year) adult (Cynthia et al., 2011). A total of 130 blood samples were collected randomly from stray dogs of Chittagong Metropolitan area. A prototype questionnaire was used to record the information like area, age, sex, etc. The process of dog handling and catching was done by humane method (also known as ‘ethological’ handling). The process is defined as causing the minimum amount of stress possible during the procedure to both the animal and the people involved (FAO, 2014). In order to achieve humane handling, the individual dog’s behaviour and the immediate environment was taken into account. Stray dogs will be caught by hand method (http://www.awbi.org/awbi-pdf/SOP) by calling or offering food to a friendly dog and gently held the dog by the scruff (loose skin on the back of the neck) of the neck with one hand, as close to the head as possible and with the other arm under the belly or around the rump. After that a face mask will be applied to control over the dog for collection of samples.
Only one biological sample (blood) was collected during this study where an individual animal was considered as a sampling unit. Smears were prepared form blood obtained from ear vein puncturing with sterile needle. Two thin blood smears were prepared by touching the coming out fresh blood and then spread by another slide. The slides were air dried and fixed by 100% methyl alcohol for 3-5 minutes (Cable, 1950). After making the slide the remaining portion of blood were kept into -20° C. All the examinations were carried out at the parasitology laboratory, Chittagong Veterinary and Animal Sciences University (CVASU). The prepared thin blood smears (Hendrix and Robinson, 2006) were stained with the Giemsa stain for 25-30 minutes. After rinsing with water, the stained blood smears were air dried and examined under binocular microscope (X100) with immersion oil for the identification of blood parasites (Soulsby, 1982; Sloss et al., 1994; Urquhart et al., 1996). Total genomic deoxyribonucleic acid (DNA) has been extracted from the whole blood samples by using PCI method (Sambrook et al., 1989). Ultimate confirmatory diagnosis was made on the basis of the PCR and DNA sequences. Although, there were different types of specific techniques the study aims to impose on the commonly used rapid diagnostic tools beside of molecular technique (PCR and Real-time PCR). Again, the study also considers only those tools which are more commonly used for wide range of organism to diagnose. The extracted DNA from all microscopic positive samples of dogs’ blood were subjected to PCR by using single set of primers containing part of 18S rRNA gene sequence. It was amplified and 422–440bp for Babesia spp (Hilpertshauser et al., 2006) (Table 1). After amplification the PCR, mixture was subjected to 1% agarose gel electrophoresis.
To detect the Babesia from samples PCR amplifications were performed at the following thermal conditions: 94°C for 2 min followed by 45 cycles of 94°C for 30 sec, 61°C for 45 sec, 72°C for 1 min and followed the final extension step at 72°C for 10 min (Hilpertshauser et al., 2006). After completing the PCR, the tubes containing PCR products were removed from the thermo cycler and stored at 4°C in a refrigerator until electrophoresis.
It is diagnostically important to determine the species, sub-species and genotype that cause canine babesiosis. For definitive diagnosis of canine babesiosis, as well as the differentiation of the species of piroplasms, a seminested PCR was performed.
For the seminested PCR, an outer primer pair (455-479F and 793-772R) was designed that would amplify an approximately ~340 bp for Bebesia gibsoni (Asian type) (AF271081, AF27182), Babesia canis, B. canis subsp. canis (AJ009795), B. canis subsp. vogeli (AJ009796) and B. canis subsp. rossi (L19079) (Birkenheuer et al., 2003a).
Then specific internal primers were designed for B. gibsoni (Asian type) (BgibAsia-F) and B. canis subsp. canis (BCC-F) that were paired with the outer reverse primer in the seminested secondary reaction to amplify 185 bp and 198 bp respectively (Birkenheuer et al., 2003a).
To detect the Babesia sp. from samples PCR amplifications were performed at the following thermal conditions: 95°C for 5 min followed by 50 cycles of 95°C for 45 sec, 58C for 45 sec, 72°C for 45 sec and followed the final extension step at 72°C for 5 min (Birkenheuer et al., 2003a).
To detect the B. gibsoni (Asian type) (BgibAsia-F) and B. canis subsp. canis (BCC-F) from samples PCR amplifications were performed at the following thermal conditions: 95°C for 5 min followed by 30 cycles of 95°C for 45 sec, 58°C for 45 sec, 72°C for 45 sec and followed the final extension step at 72°C for 5 min (Birkenheuer et al., 2003a).
1% agarose gel was made by using 0.5 g agarose powder and 50ml TAE buffer with ethidium bromide. The DNA amplicons were visualized using 4 μl of the final PCR product and 2 μl standard 100bp DNA markers (Invitrogen) at 120 V/100mA for 30 min. Gels were photographed using a gel documentation system Positive or negative amplifications were evaluated as presence or absence of visible bands on agarose gels under UV light (O’Dwyer et al., 2009).
The PCR products of representative strains were purified with the PCR Clean-up system kit (FavorPrepTM DNA purification Mini Kit) and this products were sequenced at ICCDR, B (Bangladesh) where commercial sequencing is routinely performed.
Table 1: Details of the primers used for PCR
|
PCR primer |
Primer name |
Sequences (5ʺ-3ʺ) |
Size (bp) |
|
Babesiosis |
bab- F bab- R |
5GTTTCTGMCCCATCAGCTTGAC-3’ 5CAAGACAAAAGTCTGCTTGAAAC-3 |
420-440 |
Table 2: Sequences for the oligonucleotide primers used in this seminested PCR
|
Primer |
Sequence (5′- 3′) |
Reaction and /or use |
|
455-479F |
GTCTTGTAATTGGAATGATGGTGAC |
Seminested PCR outer forward primer |
|
793-772R |
ATGCCCCCAACCGTTCCTATTA |
Seminested PCR outer reverse primer |
|
BgibAsia-F |
ACTCGGCTACTTGCCTTGTC |
Seminested PCR B. gibsoni (Asian type) forward primer |
|
BCC-F |
TGCGTTGACGGTTTGACC |
Seminested PCR B. canis subsp. canis forward primer |
Table 3: Overall prevalence of Babesia spp. in microscopic and PCR techniques
|
Techniques |
Total No. of sample (n=130) |
|
|
No. of positive sample |
Prevalence (%) |
|
|
Microscopic |
9 |
6.92 |
|
PCR |
6 |
4.61 |
The sequenced product was analyzed using the program (Chromas Lite version 2.1, 2012, Technelysium Pty Ltd, South Brisbane, Queensland, Australia) and was compared to sequence data available from GenBankTM, using the BLAST 2.1 program (http://www.ncbi.nlm.nih.gov/BLAST/).
Phylogenetic Analysis
The partial sequences of the 18S rRNA genes for B. gibsoni, B.canis were retrieved from the GenBank database and aligned with the sequence CVASU/BD/Babesia (KR349267) by using the program MEGA version 6.0 (Tamura et al., 2013). The tree was computed as neighbor joining tree by Tamura-Nei, 93 model and the replication cycle was 1000. The trees were statistically evaluated by using bootstrap methods.
Statistical Analysis
The obtained information was imported, stored and coded accordingly using Microsoft Excel-2007 to STATA/IC-11.0 (Stata Corporation College Station) for analysis. The result were expressed in percentage with P-value for Chi-Square Test. Significance was determined when P<0.05.
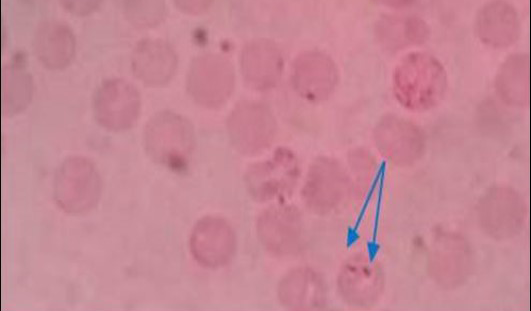
Figure 1: Pear shaped piroplasm inside RBC (x100)
Results
Prevalence of the Disease
All the samples were screened by microscopy for identification of piroplasms in Giemsa’s stain technique. Microscopic examination found a total of 9 positive (6.92%) samples. The piroplasms were seen as pyriform (pear) shape inside the red blood cells (RBC) (Figure 1). Again the samples were run by PCR using 18S rRNA gene and found 6 positive samples (Figure 2).
Table 4 shows that the prevalence of Babesia spp. in adult dog (10.11%) is higher than young and there is a significant difference between the two age groups in aspect
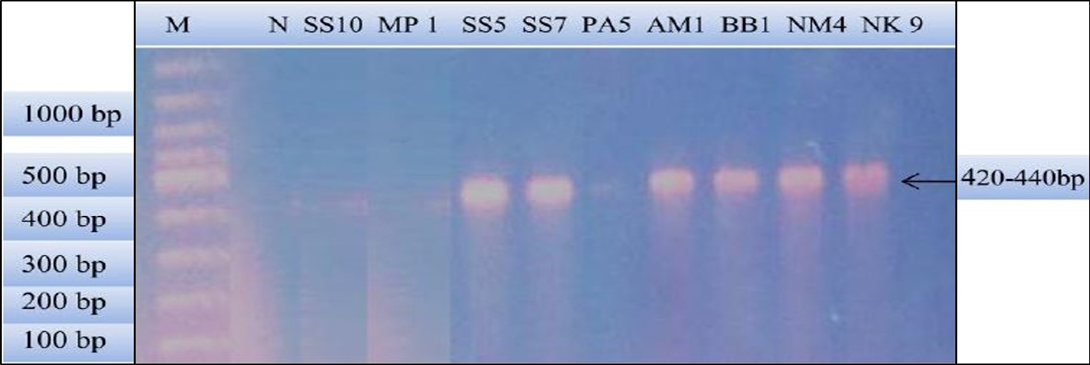
Figure 2: PCR products amplified using bab-F and bab-R specific primers: Lane M) 100 bp DNA ladder (Invitrogen); Lane N) Negative control; Lane 2, 3 and 6) Negative samples; Lane 4, 5, 7, 8 and 9) Positive samples
Table 4: Relationship of age and sex to the prevalence of Babesia spp. in stray dog
|
Variables |
N |
Microscopic examination |
Confirmation of Babesia spp. DNA by 18S rRNA gene PCR |
|||||
|
No. of positive |
% |
P value |
Microscopic positive |
PCR positive |
P value |
|||
|
Age |
Adult |
89 |
9 |
10.11 |
0.03* |
9 |
6 |
No value |
|
Young |
41 |
0 |
0.00 |
0 |
0 |
|||
|
Sex |
Male |
59 |
8 |
11.94 |
0.02* |
8 |
6 |
0.45 |
|
Female |
62 |
1 |
1.59 |
1 |
0 |
|||
N: Total number; %: percentage; *: p<0.05
of microscopic examination. Again, there is a significant difference within the two sex groups. Male dogs (11.94%) have more prevalence than female.
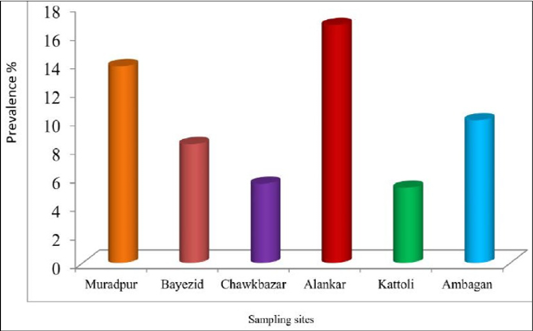
There was no statistically significant difference among the sampling sites for the prevalence of babesiosis in dog. The prevalence of babesiosis in stray dog was represented in Figure 3. The prevalence was highest (16.67%) in the samples of Alankar and lowest (5.26%) in the samples of Kattoli (Figure 3). But, there was no significant difference in the prevalence of babesiosis among the sampling sites.
Identification and Characterization of Babesia sp. by Seminested PCR
Babesia sp. was identified and characterized from the PCR positive samples run by ‘Babesiosis primer’ contained the sequences of 18S rRNA gene. For species identification and characterization a seminested PCR performed. During the primary reaction of the seminested PCR, a ~340 bp product was amplified. This result confirmed that the positive samples belong to Babesia gibsoni (Asia genotype), B. canis subsp. vogeli, B. canis subsp. canis or B. canis subsp. rossi.
During the secondary seminested reaction, the test was able to differentiate Babesia gibsoni (Asia genotype), B. canis subsp. vogeli, B. canis subsp. canis and B. canis subsp. rossi when the specific internal primers were paired with the reverse primer. In the secondary seminested reaction I have used BgibAsia-F and BCC-F as internal primers and 793-772R as reverse primer. In the second round of seminested PCR, there were six (6) positive samples by the pairing of BgibAsia-F and 793-772R (Figure 5).
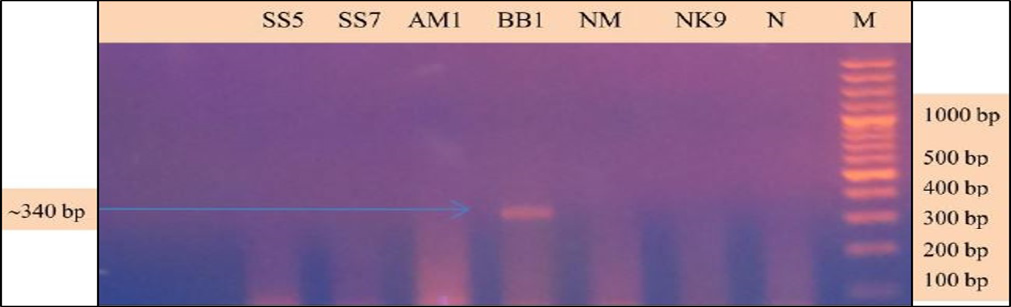
Figure 4: PCR products amplified using 455-479F and 793-772R specific primers: Lane M) 100 bp DNA ladder (Invitrogen); Lane N) Negative control; Lane 2, 3, 5, 6 and 7) Negative samples; Lane 4) Positive samples
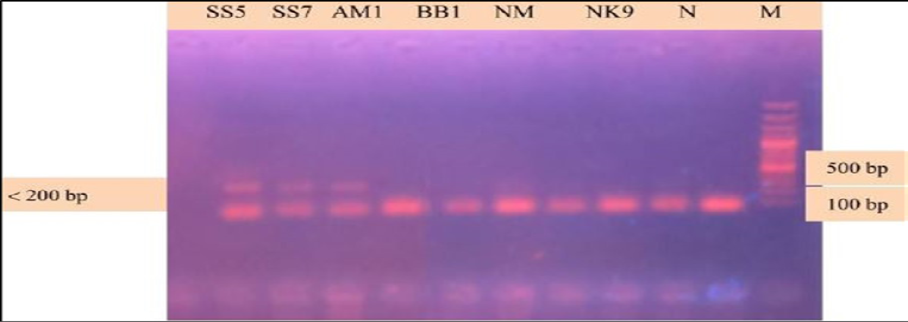
Figure 5: PCR products amplified using Bgib Asia-F and 793-772R specific primers: Lane M) 100 bp DNA ladder (Invitrogen); Lane N) Negative control; Lane 2, 3, 4, 5 and 6) Negative samples; Lane 7, 8 and 9) Positive samples
Identification and Characterization of Babesia sp. by Sequencing of PCR Product
After sequencing of the representative PCR product, the quality of the sequence was assessed manually for each nucleotide. The sequence data was used to conduct non-redundant BLASTN analysis to characterize the Babesia sp. and nucleotide sequence of the sample shown 95% identity with the other Bangladeshi Babesia gibsoni isolate BD 37 (LC008285) and Kolkata isolate (KJ142323).
Phylogenic Analysis
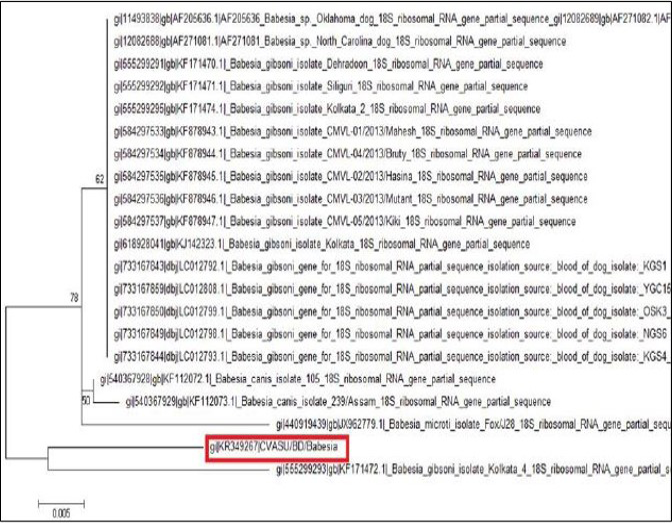
The tree was computed by using neighbor joining method of the MEGA program. Numbers at the nodes indicate the bootstrapping support for each internal branch. Scale bar indicates an evolutionary distance of 0.005 nucleotides per position in the sequence. Vertical distances are for clarity only. The GenBank accession numbers and names of the genes for the sequences were used in the analysis. The phylogenetic tree shows our isolate is most closely related to but distinct from Kolkata isolate. CVASU/BD /Babesia and Kolkata isolate forms clade (Figure 6).
Discussion
The overall prevalence of babesiosis in the current study by examination of Giemsa stained peripheral blood smears was higher than (Singh et al., 2012; Jalali et al., 2013; El-Dakhly et al., 2014). They found 5.83%, 3.75% and 1.6% in India, Iran and Japan, respectively. The higher prevalence of the disease in the present study was due to the variation of the study area or availability of the vectors. Higher prevalence of the babesiosis in the study population suggested a continuous challenge of such infection in those areas. Again the prevalence of the babesiosis in present study by microscopy was lower than (Ahmad et al., 2007; Amuta et al., 2010; Polak et al., 2012; Laummaunwai et al., 2014; Singh et al., 2014). They found 12.49%, 10.2%, 12.06%, 13.2% and 7.74% in Pakistan, Nigeria, India, Thailand and Northwest India, respectively. The lower prevalence of babesiosis in the current study might be due to random sampling rather than selection of clinically susceptible dog. However, variation in geo-climatic condition and exposure of vectors might add to the erratic prevalence of blood parasitic diseases (Rahman, 2014). The prevalence of babesiosis by PCR technique is lower than the previous reports published in Bangladesh (Talukder et al., 2013; Terao et al., 2015). They found 38.2% and 30% prevalence, respectively. The lower prevalence in the present study might be due to random sampling.
Sexual category of animals also has influences in the occurrence of vector borne parasitic diseases. In present study revealed that male dogs carrying more prevalence of babesiosis than females, which suggested that bite wounds or blood transmission during fighting contact as possible routes of transmission for B. gibsoni. Male dogs most commonly associated with dog fighting or perhaps it may be due to the aggressive behavior of males, against other males especially during mating period to be dominant and therefore they get wounded and infected, or another possibility was the hormonal status of males that may lead to high infection (Bashir, 2008; Salem and Farag, 2014). On the other hand, some reports hinted that female dogs are more prone to babesiosis than males (Singh et al., 2012, 2014). However, there is no difference in sex susceptibility between males and females (Martinod et al., 1986).
Age also influences the occurrence of haemoprotozoan diseases. In current study, higher susceptibility of adult dog to babesiosis was found consistent with some findings (Hornok et al., 2006; Salem and Farag, 2014). They reported, Babesia infection has been known to rise with the age, reaching its peak between the age of 3 and 5 years. Again B. canis was recorded only from the dogs above 1 year of age and the results are similar to earlier reports (Singh et al., 2012). Another study indicated that dogs from day 1 – 2 years of age were significantly more likely to test positive for the presence of Babesia spp. than the dogs of the other ages and B. gibsoni is transmitted transplacentally and not by the transmammary route (Fukumoto et al., 2005; Bashir, 2008). Whereas, it has also been reported that age do not have any influence on the animals’ susceptibility to the disease (Martinod et al., 1986).
For several reasons, a definitive diagnosis of canine babesiosis can be difficult to achieve in the clinical background. Light microscopic examination cannot consistently differentiate species or subspecies. The lack of standardized serologic assays, the presence of cross-reactive antibodies and recent changes in the geographic ranges of several canine piroplasms have also further complicated the diagnosis of babesiosis in dogs. In the present study we detected Babesia organism via PCR, however, the cost, equipment, and time may be a major limitation for the use of PCR in clinic-based practice (Irwin, 2007; Solano-Gallego et al., 2008). Again, in the present study, we describe a seminested PCR, which is specific for the diagnosis and differentiation of Babesia gibsoni (Asia genotype), B. canis subsp. vogeli, B. canis subsp. canis or B. canis subsp. rossi. In the seminested PCR we found Babesia gibsoni (Asian type) which was consistent with the earlier report published in Bangladesh (Talukder et al., 2013; Terao et al., 2015).
Amplification of the partial 18S rRNA gene, by nusing generic protozoan primers and the Babesia DNA extracted from the dog blood, yielded a specific product of approximately 349 base pairs. Sequence analysis of this gene gives an intention and specific means of species identification and phylogenetic classification. BLAST (available from: http://www.ncbi.nlm.nih.gov/BLAST/) search showed that the sequence, although clearly from a Babesia gibsoni, was not identical to any complete 18S rRNA sequences in the GenBank database. The phylogenetic tree shows our isolate (K349267) is most closely related to but distinct from Kolkata isolate (KJ142323). The DNA sequences of the 18S rRNA gene were identical for the Babesia organism from the two dogs, which indicate that they were infected with the same organism Babesia gibsoni. Each of the organisms was sequenced in a different country, which indicates that the findings were not artifactual. Again, phylogenetic tree shows that these two isolates form clade which indicates geographical location may have some influence on the distribution of the parasite.
Conclusion
The study was performed aiming to determine the prevalence of babesiosis in stray dog and standardization of PCR-based molecular diagnosis and identification of babesiosis. Canine babesiosis was strongly associated with the age and sex of the animals where adult male dogs were more susceptible than others. The present study also revealed that PCR is more specific than microscopic examination. Babesia gibsoni was the dominant organism for canine babesiosis in this part of Bangladesh which was established by a seminested PCR and phylogeny through gene sequencing. Finally, we recommended further studies focusing identification of tick vectors along with molecular detection of organisms from tick for taking further control strategies in the study areas.
Acknowledgement
We greatly acknowledge the support provided by UGC-HEQEP Round –II sub project (CP: 2180). We are also thankful to Department of Pathology and Parasitology of CVASU for their support and inspiration during the study period.
Conflict of interest
The authors declare that they have no conflict of interest.
Authors’ contributions
NEA is the main and corresponding author, carried out the study, analyzed the result, drafted the manuscript and submitted. SI helped in sample collection and made the experimental design, MT participated in sequence alignment, TMR helped in sample collection, EB and AM helped in molecular study and MAH supervised the whole study. All authors read and approved the final manuscript.
References