Advances in Animal and Veterinary Sciences
Research Article
First Report of Molecular Identification and Characterization of Theileria spp. from Water Buffaloes (Bubalus bubalis) in Egypt
Amira Adel Taha Al-Hosary1*, Laila Ahmed1, Ulrike Seitzer2
1Department of Animal Medicine (Infectious Diseases), Faculty of Veterinary Medicine, Assiut University, Assiut, P. O. Box 71526, Egypt; 2Division of Veterinary Infection Biology and Immunology, Research Center Borstel, Parkallee 22, 23845, Borstel, Germany.
Abstract | In the present study, 37 buffalo from different localities in Upper Egypt were sampled and examined for Theileria spp. infection using Giemsa blood smears, conventional and nested polymerase chain reaction. The analysis targeted the 18S srDNA gene followed by sequencing and phylogenetic analysis using neighbor-joining (NJ) method. The study came up with the conclusion that Egyptian buffalos (Bubalus bubalis) infected with three Theileria (T. uilenbergi (13.5%), T. lestoquardi (2.7%) and T. ovis (2.7%)) small ruminants species rather than Theileria annulata (32.4%).
Keywords | Bubalus bubalis, Theileria spp., Theileria annulata, T. uilenbergi, T. lestoquardi, T. ovis
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | September 13, 2015; Revised | October 06, 2015; Accepted | October 07, 2015; Published | November 04, 2015
*Correspondence | Amira Adel Taha AL-Hosary, Assiut University, Assiut, Egypt; E-mail: amiraelhosary@ yahoo.com
Citation | Al-Hosary AAT, Ahmed L, Seitzer U (2015). First report of molecular identification and characterization of Theileria spp. from water buffaloes (Bubalus bubalis) in Egypt. Adv. Anim. Vet. Sci. 3(12): 629-633.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.12.629.633
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Al-Hosary et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bovine tropical theileriosis (Theileria annulata infection) is a destructive disease that affects the livestock productivity in several enzootic countries. In Egypt, this disease affects both cattle and buffalos and is considered as a serious health problem especially in Upper Egypt which includes eight governorates along the Nile River from Cairo in the North to Aswan in the South (EL-Nahrawy, 2011). Buffalos are considered as the primary dairy animal in Egypt; the average number of water buffaloes (Bubalus bubalis) population in Egypt is around six millions including 42% dairy buffalo cows. Buffalos’ milk represents 80% of the whole Egyptian milk production. Due to its high quality and high fat percentage it is the preferred milk source in Egypt. Thanks to their high conversion food rate, buffalos are preferred to cattle in poor food resources regions (Ibrahim, 2012). Due to their high rusticity, buffaloes act as reservoirs for several pathogens including tropical theileriosis especially in mixed farms (Michel and Bengis, 2012). Buffalos are natural hosts of Theileria annulata but remain frequently carriers due to their ability to control the infection by cellular and humeral mediated immune response (Stagg et al., 1983; Baldwin et al., 1986). The development of molecular methods like polymerase chain reaction (PCR) provides a highly sensitive and specific diagnosis tool in both clinically infected and carrier animals (AL-Hosary et al., 2009; Abdel-Rady et al., 2010). The present study aimed to carry out a molecular characterization and phylogenetic analysis of Theileria spp. parasites infecting buffaloes in Egypt.
Materials and methods
Animals and Sampling
A total number of thirty-seven buffalo cows (Bubalus bubalis) belonging to different localities in Upper Egypt were sampled during August 2010 to August 2012. The age of these animals was ranged from one year to more than 5 years of both sexes. Two blood samples were collected, one from ear vein and used for preparation of Giemsa strained blood smear, the second was collected from jugular vein on EDTA vacutainer tubes and stored at -20°C till used (Charles, 2002).
Conventional Detection
Blood smears were prepared directly after sampling, fixed with methanol, stained with 10% Giemsa stain and
Table 1: Results of different diagnostic assays according different isolates
|
Theileria spp |
Blood smear |
Standard PCR |
Nested PCR |
|||
|
Positive |
Prevalence ± SD |
Positive |
Prevalence ± SD |
Positive |
Prevalence ± SD |
|
|
T. annulata |
3 |
8.1 ± 5.2 |
3 |
8.1 ± 5.2 |
12 |
32.4 ± 5.2 |
|
T. Uilenbergi |
0 |
0 ± 2.9 |
0 |
0 ± 2.9 |
5 |
13.5 ± 2.9 |
|
T. ovis |
0 |
0 ± 0.58 |
0 |
0 ± 0.58 |
1 |
2.7 ± 0.58 |
|
T. lestoquardi |
0 |
0 ± 0.58 |
0 |
0 ± 0.58 |
1 |
2.7 ± 0.58 |
examined under microscope at x1000 magnification with immersion oil.
Molecular Detection
DNA Extraction
DNA extraction from whole blood was performed with QIAamp blood kit (Qiagen, Ltd, UK) according to manufacturer’s instructions; DNA was stored at -20ºC till use.
PCR and Nested-PCR
The V4 gene 18S rRNA hypervariable region was amplified by using forward (RLB-F 5’- GAC ACA GGG AGG TAG TGA CAA G-3’), Reverse (5’- CTA AGA ATT TCA CCT CTG ACA GT-3) and nested forward (5’-GAC AAG AAA TAA CAA TAC RGG GC-3’) primers. In each PCR run, positive and negative control tubes were used, consisting of genomic Theileria annulata DNA and molecular grade water, respectively. Reaction was performed in a total volume of 35 µl containing 3.8 µl 10x PCR buffer, 0.8 µl of each dNTP, 1.8 µl of each forward and reverse primers, 0.18 µl Super Taq polymerase, 28 µl molecular grade water and 2 µl DNA template. Cycling condition was set according to Gubbels et al. (1999).
Cloning and DNA Sequencing
PCR products were ligated on pDrive Cloning vector using Qiagen PCR Cloning Kit according to manufacturer’s instructions (Qiagen, Ltd, UK). The vector then was transformed into competent cells by using XL1-Blue competent cells kits according to manufacturer’s instructions (Stratagene, Agilent Technologies Division). The competent cells allowed to grown on LB plates for 24 hours at 37°C then followed by blue/white colony selection. High copy plasmid DNA was purified from 7 ml bacterial lysates, it was performed according to commercial kits manufacturer’s instructions (Analytikjena/Biometra Life Science unlimited, innuPREP Plasmid Mini Kit, Germany). The amplicons were sequenced in both directions and subjected to BLAST similarity searches.
Phylogenetic Analysis
Phylogenetic analysis was performed after aligning sequences by neighbor joining method using the Mega 6.0 program. This method has desirable statistical properties and is known to produce trees as accurate as more computationally intensive. Significance of branching within a phylogenetic tree was tested by BOOTSTRAP TREE program. The phylogenetic trees were constructed with Kimura two parameters distance and the nodes were tested for robustness by 1,000 bootstrap replications (Kimura, 1980).
Results
Blood smears T. annulata infection prevalence was estimated to 8.1% (3/37). Standard and nested PCR prevalence were 8.1% (3/37) and 51.3% (3/37) respectively (Table 1, Figures 1 and 2).
Sequences alignment and phylogenetic analysis by NJ method revealed that the partial sequences of the 18S srDNA gene isolated from of the Egyptian water buffaloes fall inside the Theileria genus clade. Theileria sequences shared 99:100% identity with other Theileria sequences. Two sequences were related to T. annulata; one was related to T. lestoquardi, another to T. ovis and four sequences were related to T. uilenbergi. All of these partial sequences were deposited in the GenBankTM databases and available under the following accession numbers: T. annulata (KF922216; KF922219), T. lestoquardi (KF771185), T. ovis (KF922218)
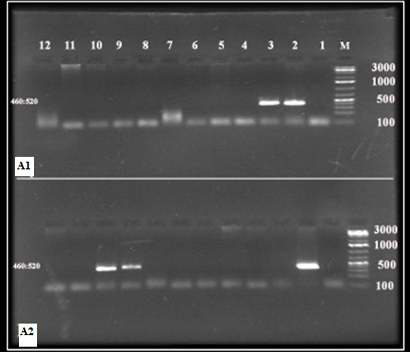
Figure 1: 18S srRNA PCR products
Lane M: DNA ladder 100 bp, (A1) Lanes 2 and 3, (A2); Lanes 2, 9 and 10: positive PCR both of them gave bands of 460 and 520 bp.
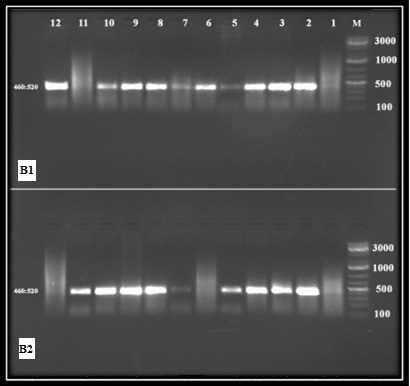
Figure 2: 18S srRNA nested RLB-PCR products
Lane M: DNA ladder 100 bp, (B1); Lanes 2, 10 and 12: (B2) 2:5 and 7:11 positive nPCRs both of them gave bands of 460 and 520 bp.
and T. uilenbergi (KF781308; KF771184; KF922215; KF922217) (Figure 3).
Discussion
The current study identified several species of Theileria rather than T. annulata that includes T. lestoquardi, T. uilenbergi and T. ovis. According to our knowledge, this is the first report of T. lestoquardi and T. uilenbergi in Egypt. These species were reported in Sudan, Tanzania, Iraq, Iran, Oman, Tunisia and China (Schnittger et al., 2000b, 2004; Shayan et al., 2011; Ahmed et al., 2013; Ranneker et al., 2013; Rjeibi et al., 2014 a, b). Several studies mentioned that buffaloes are more resistant to Theileria spp. infections than cattle and this could be attributed to the ability of its innate immunity and cell-mediated immunity to control the infection. In addition, the protozoan parasite has low affinity to the buffaloes’ cells (Stagg et al., 1983; Baldwin et al., 1986). This explains why some buffaloes show no clinical signs when infected and remain subclinically infected or carrier (Michel and Bengis, 2012). The role of buffaloes as a reservoir is to be investigated since they could activate parasite mutations rate and provide modified drug resistant species for cattle and small ruminants. The Occurrence of T. annulata was expected since it is the only Theileria species infecting buffalos in Egypt (Osman and AL-Gaabary, 2007; El-Deeb and Younis, 2009; Mohamed et al., 2011; EL-Sify et al., 2015). The three previously mentioned Theileria species are infecting sheep and should arise the status of these species as emerging. T. lestoquardi and T. annulata have the same vector: Hyalomma anatolicum and their
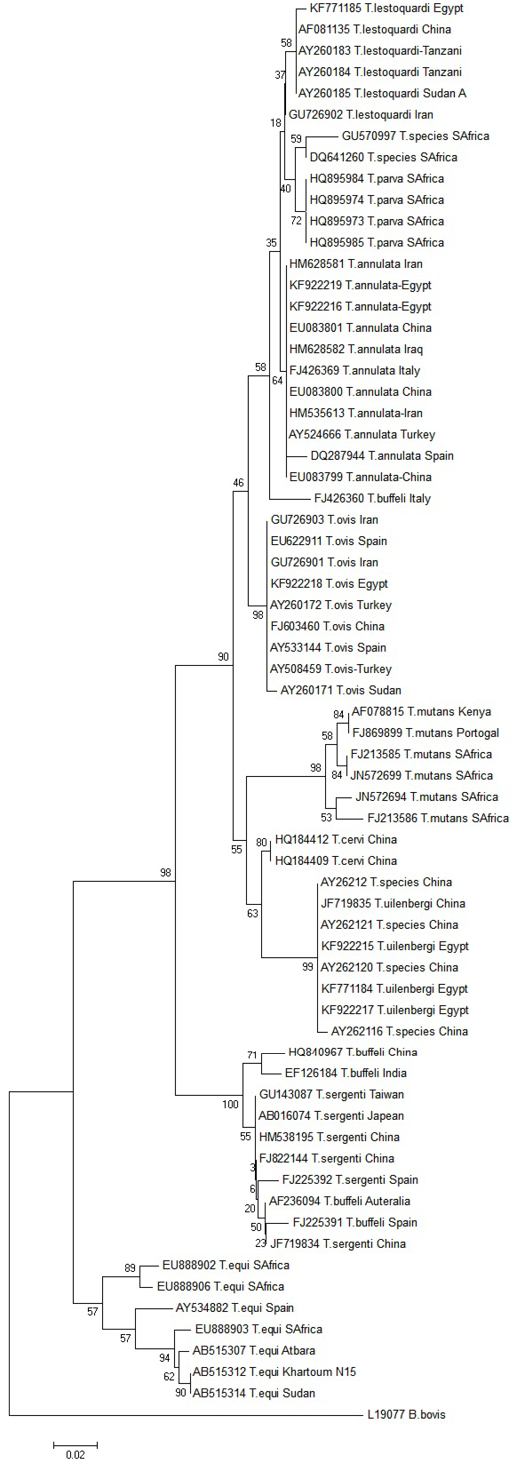
Figure 3: Phylogenetic tree of the 18S srRNA gene
geographic distribution tend to overlap in Sudan (Tahaa et al., 2013). Therefore, previous study suggested that T. annulata evolved relatively recently from a common ancestor with T. lestoquardi (Leemans et al., 1997; Tahaa et al., 2013). T. uilenbergi is genetically different from T. annulata and T. lestoquardi, the identity between them was the lowest when compared to other Theileria species. T. uilenbergi is still undergone more extended individual evolution as mentioned by Schnittger et al. (2000a, b). Theileria ovis, a small ruminant’s parasite, is reported for the first time in buffaloes from Egypt. These infections by unusual pathogens may be due the fact that buffaloes are housed under poor hygienic conditions, neglected tick control programs and cohousing with small ruminants. The prevalence of T. annulata, T. uilenbergi, T. lestoquardi and Theileria ovis were 32.4; 13.5; 2.7 and 2.7%, respectively. These results are the first step to start a comprehensive epidemiological study of these parasites in buffaloes to know their role in different Theileria species cycles.
Acknowledgement
This work was supported by DFG project “Molecular epidemiology network for promotion and support of delivery of life vaccines against Theileria parva and Theileria annulata infection in Eastern and Northern Africa” (DFG project SE862/2-1).
Conflict of interest
The authors declare that they have no conflict of interests.
Author’s contribution
Amira A. T. Al-Hosary, analysed the data, interpreted and performed the experiment and wrote the paper. Laila S. Ahmed, revised the paper and conceptual advice. Ulrike Seitzer, designed the experiment and provied technical support.
References






