Advances in Animal and Veterinary Sciences
Review Article
An Overview of Heterologous Expression Host Systems for the Production of Recombinant Proteins
Amitha Reena Gomes1, Sonnahallipura Munivenkatappa Byregowda1, Belamaranahally Muniveerappa Veeregowda2, Vinayagamurthy Balamurugan3*
1Institute of Animal Health and Veterinary Biologicals, KVAFSU, Hebbal, Bengaluru-560024, Karnataka, India; 2Department of Microbiology, Veterinary College, KVAFSU, Hebbal, Bengaluru-560024, Karnataka, India; 3Indian Council of Agricultural Research-National Institute of Veterinary Epidemiology and Disease Informatics (ICAR-NIVEDI), Post Box No. 6450, Yelahanka, Bengaluru-560064, Karnataka, India.
Abstract | With the advent of recombinant DNA technology, cloning and expression of numerous mammalian genes in different systems have been explored to produce many therapeutics and vaccines for human and animals in the form of recombinant proteins. But selection of the suitable expression system depends on productivity, bioactivity, purpose and physicochemical characteristics of the protein of interest. However, large scale production of recombinant proteins is still an art in spite of increased qualitative and quantitative demand for these proteins. Researchers are constantly challenged to improve and optimise the existing expression systems, and also to develop novel approaches to face the demands of producing the complex proteins. Here, we concisely review the most frequently used conventional and alternative host systems, with their unique features, along with advantages and disadvantages and their potential applications for the production of recombinant products.
Keywords | Expression host systems, Heterologous proteins, Production, Prokaryotic, Eukaryotic
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 07, 2016; Accepted | June 12, 2016; Published | July 01, 2016
*Correspondence | Vinayagamurthy Balamurugan, Indian Council of Agricultural Research-National Institute of Veterinary Epidemiology and Disease Informatics (ICAR-NIVEDI), Post Box No. 6450, Yelahanka, Bengaluru-560064, Karnataka, India; Email: balavirol@gmail.com; b.vinayagamurthy@icar.gov.in
Citation | Gomes AR, Byregowda SM, Veeregowda BM, Balamurugan V (2016). An overview of heterologous expression host systems for the production of recombinant proteins. Adv. Anim. Vet. Sci. 4(7): 346-356.
DOI | Http://dx.doi.org/10.14737/journal.aavs/2016/4.7.346.356
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Gomes et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Expression host systems are employed for the expression of recombinant proteins both for therapy and research. The need for novel expression host platforms increases with the number of gene-targets for various industrial productions. Platforms already in use, range from bacteria (Baneyx, 1999), yeast (Cregg et al., 2000; Malys et al., 2011) and filamentous fungi (Visser Hans, 2011) to cells of higher eukaryotes (Altmann et al., 1999; Kost and Condreay, 1999). While choosing among these cells, both economic and qualitative aspects have to be considered. Industrial production depends on the use of inexpensive media components and high anticipated economic yield from the product. The quality of the product is very essential, especially in medicine where production of human pharmaceuticals is regulated under strict safety aspects (Gellissen, 2005). Thus, suitable host system needs to be selected depending on the purpose. Use of recombinant DNA (rDNA) technology for cloning and subsequent expression of particular gene of interest of virus/ microbes in an appropriate system like bacterial / mammalian / insect / yeast / plant expression systems will circumvent the difficulties associated with the production of large quantities of vaccine or diagnostic agents (Balamurugan et al., 2006). Such a bio-engineered protein can be obtained in large amounts in a pure and native form rDNA technology has necessary tools to produce desired viral / bacterial proteins in a native form. The advent of rDNA technology and its application in the industry has brought about a rapid growth of biotechnology companies for the production of the rDNA products in human and animal healthcare.
Prokaryotic Expression Systems
Bacterial expression system is widely used for the expression of rDNA products. They offer several advantages viz., high level of recombinant protein expression, rapid cell multiplication and simple media requirement. However, there are some limitations such as intracellular accumulation of heterologous proteins, improper folding of the peptide, lack of post-transcriptional modifications, the potential for product degradation due to trace of protease impurities and production of endotoxin (Balamurugan et al., 2006). Prokaryotic platforms are widely used to produce recombinant proteins as these can be easily manipulated genetically by well-known methods and cultivated in low costing medium. Among Gram-negative bacteria, E. coli is extensively used, while Bacillus subtilis is a well-known Gram-positive producer of recombinant proteins (Watson et al., 1992). Among the several challenges protein insolubility and low expression levels (especially mammalian proteins) or post-translational modifications are some of the intriguing problems while using these systems (Gellissen, 2005).
Escherichia coli
E. coli is a typical prokaryotic expression system and one of the most attractive heterologous protein producer. The expression of proteins in this system is the easiest, quickest and cheapest. To date reformed E. coli is the extensively used cellular host for foreign protein expression because of its rapid growth rate which is as short as 20-30 minutes (Snustad and Simmons, 2010), capacity for continuous fermentation and relatively low cost. There are many commercial and non-commercial expression vectors available with different N and C terminal tags and many different strains are being optimized for special applications.
There are also problems related to the use of E. coli as production host. These problems can be grouped into two categories: those that are due to the sequence of the gene of interest and those that are due to the limitations of E. coli as a host (Brown et al., 2006). In the first category again there are three ways in which the nucleotide sequence might prevent efficient expression of a foreign gene. Firstly, the foreign gene may contain introns which would be a major problem since E. coli genes do not contain introns and so the bacterium does not possess necessary machinery for removing introns from transcripts. Secondly, the foreign gene might contain sequences which act as termination signals in E. coli. These sequences are perfectly innocuous in the normal host cell, but in the bacterium it results in the premature termination and a loss of gene expression. Thirdly, a problem with codon bias of the gene making E. coli not an ideal host for translation. Although all organisms use the same genetic code, each organism has a bias towards preferred codons. This bias reflects the efficiency with which the tRNA molecules are able to recognize the different codons. If a cloned gene contains a high proportion of unfavoured codons, the host cell’s tRNAs may encounter difficulties in translating the gene, reducing the amount of protein that is synthesized (Brown et al., 2006).
These problems can be solved, with necessary manipulations although it is time consuming. If the gene contains introns then its complementary DNA (cDNA) prepared from the mRNA and so lacking introns might be used as an alternative. In vitro mutagenesis can be employed to change the sequences of possible terminators and to replace un-favoured codons with those preferred by E. coli. An alternative with genes that are less than 1 kb in length is to make an artificial version. This involves synthesizing a set of overlapping oligonucleotides that are ligated together, the sequences of the oligonucleotides are designed to ensure that the resulting gene contains preferred E. coli codons and terminators are absent (Brown et al., 2006).
In the second category, there are three major limitations in which E. coli may prevent expression of a foreign gene. Firstly, E. coli might not process the recombinant proteins correctly especially with respect to post translational modifications like N and O linked glycosylation, fatty acid acylation, phosphorylation and disulfide-bond formation that are required for proper folding of the secondary, tertiary and quaternary structures and for the functional characteristics of the protein of interest. Glycosylation is extremely uncommon in bacteria and recombinant proteins synthesised in E. coli are never glycosylated correctly. This can affect the bioactivity, function, structure, solubility, stability, half-life, protease resistance and compartmentalization of the functional proteins (Jung and Williams, 1997). Secondly, E. coli might not correctly fold the recombinant proteins and is unable to synthesise disulphide bonds present in mammalian proteins. In such case, protein may become insoluble and forms an inclusion body within the bacterium (Leonhartsberger, 2006). Although proteins can be recovered from such state, converting them to correct form is difficult and more often they remain inactive. Recently it has been identified that the catalytic domain of a cellulase (Cel-CD) from Bacillus subtilis can be secreted into the medium from recombinant E. coli in large quantities without its native signal peptide. It has also been proved that the N-terminal sequence of the full length Cel-CD played a crucial role in transportation through both inner and outer membranes. By subcellular location analysis, it is identified that the secretion is a two-step process via the SecB-dependent pathway through the inner membrane and an unknown pathway through the outer membrane (Gao et al., 2016). Thirdly, E. coli may degrade the recombinant protein. Unlike sequence problems, these problems can be circumvented by using mutant strains of E. coli.
Accumulation of endotoxins (LPS), pyrogenic to humans and animals, is yet another problem in the production of therapeutic proteins in E. coli, besides instability of is plasmid or mRNA (Terpe, 2006). Proteins expressed in E. coli also retain their amino terminal methionine, thus affecting the stability and in turn immunogenicity of proteins (Daly and Hearn, 2005). Further, expression of mammalian proteins in E. coli remains difficult and often results in inactive aggregates because the recombinant proteins do not fold properly. However attempts have been made to produce soluble prion proteins in E. coli (Abskharon et al., 2012).
Bacillus subtilis
It is an alternative to the E. coli expression system. B. subtilis, also known as hay bacillus or grass bacillus, is a Gram positive, catalase positive bacterium, found in soil and the gastrointestinal tract of ruminants and humans. It can secrete degradative enzymes or antibiotics, produce spores and can become competent for genetic transformation (Maamar and Dubnau, 2005).
It is a non-pathogenic and a GRAS (Generally regarded as safe) organism. The major advantage of B. subtlis is that it does not produce LPS, which may otherwise cause degenerative disorders in humans and animals. B. subtilis can also be transformed readily with many bacteriophages and plasmids due to its genetic characteristics. It is capable of secreting functional extracellular proteins directly into the culture medium, facilitating further purification steps. It has no significant bias in codon usage which is considered as an added advantage. Processes such as transcription, translation, protein folding, secretion mechanisms, genentic manipulations and large scale fermentation has been very well acquainted with this organism. The organism is also useful for the construction of metagenomic libraries (Luan et al., 2014).
Like any other system this also is not without drawbacks and they include:-
- 1. Production of extracellular proteases which recognize and degrade heterologous proteins, but can be solved to a larger extent by construction of protease deficient strains (Van Schaik and Abee, 2005; Westers et al., 2005; Servant et al., 2005).
- 2. Instability of plasmids which can be again overcome by introducing plasmids using a theta mode of replication (Janniere et al., 1990; Titok et al., 2005).
- 3. Reduced or non-expression of the protein of interest.
Bacillus megaterium
Bacillus megaterium has been used for the production and secretion of recombinant proteins for many years now. Plasmids with different inducible promoter systems, with different compatible origins, with small tags for protein purification and with various specific signals for protein secretion were combined with genetically improved host strains. Along with the overproduction of individual proteins the organism is also used for the simultaneous coproduction of up to 14 recombinant proteins, multiple subsequently interacting or forming protein complexes (Biedendieck, 2016).
Lactococcus lactis
Of late, Lactococcus lactis is widely used in biotechnology for large-scale production of heterologous proteins (Le Loir et al., 2005). During the past two decades, remarkable progress has been made towards the development of genetic engineering tools and the molecular characterization of L. lactis (Wegmann et al., 2007; Allain et al., 2016).
Ralstonia eutropha
Ralstonia eutropha formerly known as Alcaligenes eutrophus is a Gram negative, facultative chemoautotrophic bacterium found in water and soil. This system overcomes some of the shortcomings of E. coli based systems, particularly the formation of inclusion bodies during high-level expression (Srinivasan et al., 2002). Gavin et al. (2004) expressed soluble, active, organophosphohydrolase (OPH), at titers greater than 10 g/L in high cell density fermentation which is approximately 100-fold greater than titers reported in E. coli in which it tends to form inclusion bodies.
Pseudomonas
It is considered as a potential alternative to E. coli. Pseudomonas fluorescens has been proven to function as a recombinant protein producer as it can be cultivated to high cell densities often producing soluble proteins, while E. coli produces insoluble protein (Squires et al., 2004). Tonje (2011) while comparing the expression of heterologous proteins P. fluorescens was found to have low stability for the expression of plasmids than P. putida which was having high plasmid stability and proved to be potential recombinant protein producer for industrial use with growth properties similar to E. coli during fed batch fermentations.
Corynebacterium
Non-pathogenic species of the Gram-positive Corynebacterium are used for the commercial production of various amino acids. The C. glutamicum is used for producing glutamate and lysine (Brinkrolf et al., 2010), components of human food, animal feed and pharmaceutical products. Expression of functionally active human epidermal growth factor has been done in C. glutamicum (Jump et al., 2006), thus demonstrating a potential for industrial-scale production of human proteins.
Eukaryotic expression systems
In theory, prokaryotic hosts can express any gene, but in practice the proteins produced do not always have the desired biological activity or stability. Further, toxic components from the bacteria may contaminate the final product. This becomes a major issue particularly when the expressed protein is intended for therapeutic use. One must ensure that the recombinant protein must be identical to natural protein in all its properties. As eukaryotic cells share many molecular, genetic and biochemical features, it can be used as an alternative to prokaryotic expression of proteins.
Yeast System
Yeast is an expression system with the highest commercial value. Among yeast, Saccharomyces cerevisiae (Baker’s yeast) is the most widely used. Further, it has been widely used as a model organism for cell function research as its biochemistry, genetics and cell biology is very well characterized to express a number of proteins including proteins to be used for vaccines, pharmaceutical products and for diagnostics (Glick et al., 2010). It was engineered to express different heterologous genes for almost 25 years (Hitzeman et al., 1981). It satisfies the economic efficiency and biosafety regulations for human applications. This system is used successfully to make hepatitis B vaccine (DiMiceli et al., 2006) and Hantavirus vaccine (Antoniukas et al., 2006). It has been used to optimize the functional yields of potential antigens for the development of subunit vaccines against a wide range of diseases caused by bacteria and viruses. Saccharomyces cerevisiae is used in the manufacture of 11 approved vaccines against hepatitis B virus and one against human papillomavirus; in both cases, the recombinant protein forms highly immunogenic virus-like particles (Bill, 2015).
Among the eukaryotic systems, yeast is unique in that it combines the advantages of both prokaryotic [high expression levels (10-100 fold higher), faster growth, easy maintenance, easy scale-up, inexpensive growth media] and eukaryotic (capacity to carry out most of the post-translational modifications like protein processing, protein folding etc.,) expression systems. Yeast such as Saccharomyces cerevisiae, Hansenulla polymorpha and P. pastoris are among the simplest eukaryotic organisms, which grow relatively quickly and are highly adaptable to large-scale production, Technical advantages in this system include site-specific integration, increase in copy number, leader sequence for the secretion of heterologous protein and post-translational modifications (Balamurugan et al., 2006).
In recent past, the methylotrophic yeasts, such as Hansenulla polymorpha, Pichia pastoris and Candidia biodini have been developed, among which P. pastoris has emerged as a powerful and inexpensive heterologous system for the production of high levels of functionally active recombinant proteins (Balamurugan et al., 2007), in addition to existing Saccharomyces. Intact protein production and secretion into the medium makes yeast could be an efficient system for production and purification of the expressed protein (Balamurugan et al., 2006).
Presently, Pichia pastoris (Zhou et al., 2006; Ahmad et al., 2014) and Schizosaccharomyces pombe (fission yeast) (Kumar and Singh, 2004; Benko et al., 2016) are being increasingly used yeast systems for heterologous protein expression besides S. cerevisiae.
The enriched endomembrane system of yeasts does allow some intracellularly synthesized proteins to be secreted into the extracellular environment. As a unicellular eukaryote, yeast can produce properly folded soluble recombinant proteins with required post-translational modifications that are essential for their functions (Daly and Hearn, 2005). The safety of the system is guaranteed, by not having endotoxins and oncogenes. Moreover, yeast cells are easier to manipulate genetically than mammalian cells and can be grown to high cell densities.
But, this “lower” eukaryote differs from its mammalian counterparts in the way it forms both N and O linked oligosaccharide structures on target proteins (Jung and Williams, 1997). Particularly, S. cerevisiae is unable to glycosylate animal proteins correctly and it may often add many sugars leading to hyper-glycosylation of proteins. Recently, advances in the glycoengineering of yeast and the expression of therapeutic glycoproteins with humanized N glycosylation structures have shown a significant promise (Wildt and Gerngross, 2005). Besides codon biasness, yeast system is an inefficient one in secreting the proteins into growth medium leading to intracellular retention making them lot more difficult to purify.
Pichia pastoris makes a good alternative with glycosylation abilities similar to those of animal cells. Even though, the sugar structures it synthesizes are not same as the animal versions but the differences are relatively trivial and does not have a significant effect on the activity of a recombinant protein.
Meanwhile, several “non-conventional” yeasts (Gellissen et al., 2005) are well established as expression systems, which include Arxula adeninivorans (Terentiev et al., 2004), Hansenula polymorpha (Kulkarni et al., 2006), Kluyveromyces lactis (Donnini et al., 2004) and Yarrowia lipolytica (Madzak et al., 2004).
Fungus System
Filamentous fungi, especially Aspergillus (Aspergillus niger and Aspergillus oryzae) and Trichoderma have been developed into expression platforms for screening and production of diverse industrial enzymes. Trichoderma reesei has tremendous capability to secrete over 100 g/L of proteins and therefore makes an excellent host system for production of high levels of therapeutic proteins at low cost (Landowski et al., 2016). Fungal-based systems have several advantages due mainly to their high-level secretion of enzymes and their decomposer lifestyle. Further, in the large-scale production of recombinant proteins of eukaryotic origin, the filamentous fungi become the system of choice due to critical processes shared in gene expression with other eukaryotic organisms. But, the complexity and relative dearth of understanding the physiology of filamentous fungi, compared to bacteria, have hindered rapid development of these organisms as highly efficient factories for the production of heterologous proteins (Su et al., 2012). More recently Myceliophthora thermophila, C1 (Visser Hans, 2011) has been developed into an expression platform for screening and production of diverse industrial enzymes. C1 shows a less viscous morphology in submerged culture, enabling the use of complex growth and production media. T. reesei strains suitable for production of therapeutic proteins by reducing the secreted protease activity have also been developed recently (Landowski et al., 2016).
Insect System
Insect cell culture systems are widely used for the production of recombinant proteins, vaccines and viral pesticides as well as in the basic research in biology. A large number of cell lines from diptera, hemiptera and lepidopteran insects have been established. High levels of heterologous gene expression are often achieved compared to other eukaryotic expression systems, particularly for intracellular proteins (Balamurugan et al., 2006). In many cases, the recombinant proteins are soluble and easily recovered from infected cells late in the infection when host proteins synthesis is diminished. Insect cell based systems especially baculovirus based systems revolutionized the recombinant protein production. Baculovirsuses have a restricted host range limited to specific invertebrate species. Being noninfectious to vertebrates, these viruses are safer to work with than most mammalian viruses. Most of the susceptible insect cell lines are not transformed with pathogenic or infectious viruses and can be cared for under minimal containment conditions. Helper cell lines or helper viruses are not
needed since the baculovirus genome contains all the genetic information needed for propagation in a variety of cell lines or larvae from different insects. Baculoviruses are usually propagated in insect cell lines derived from the fall armyworm, Spodoptera frugiperda or from the cabbage looper, Trichoplusia ni. Commonly used and
commercially available insect cell lines are Sf-9, Sf-21 and high five tTrichoplusia ni.). Prolific cell lines are available which grow well in suspension cultures, permitting the production of recombinant proteins in large-scale bioreactors. The recombinant proteins can be produced in insect cell lines as well as in insects. This recombinant virus can be prepared by cloning any DNA insert coding protein of desire under polyhedron growth promoter or Pro promoter. The recombinant viruses are selected by their inability to induce inclusion body formation 72 hrs after infection of the cell lines or infecting insects for the production of recombinant protein. The production level in this system is very high and if the recombinant virus is prepared for infecting insects then the production of protein is very cheap. However, there is one drawback with this system that glycosylation of protein in insect cells is different from mammalian cells which leads to improper maintenance of epitopes in the target protein (Balamurugan et al., 2006).
Baculoviruses are insect pathogens that regulate insect populations in nature and are being successfully used to control insect pests. Typical property of very late gene expression makes them highly suitable as vectors for foreign gene expression. Two baculoviruses are broadly applied in biotechnology as vectors to produce recombinant proteins in insect cells: Autographa californica multiple nucleopolyhedrovirus (AcMNPV) and to a lesser extent Bombyx mori (Bm) NPV. An additional advantage is that they have a limited insect host range and hence safe for vertebrates. Insect cells can grow in serum-free media and the cultures can easily be scaled up (Smagghe et al., 2009; van Oers and Lynn, 2010). The lepidopteran insect cells are also free of human pathogens. The proteins produced in the baculovirus-expression system are used for functional studies, vaccine preparations or diagnostics.
The first vaccine produced in the baculovirus expression system, which was commercialized and approved, is directed against classical swine fever or hog cholera (Intervet) and is accompanied by a serological test (Bouma et al., 1999; van Rijn et al., 1999) thus allowing the differentiation between infected and vaccinated animals. The system is valuable for the production of proteins for structural studies and G protein-coupled receptors. It is also used for production of the human papilloma virus vaccine, Cervarix, the first FDA approved insect cell produced product and FluBlok, a vaccine based on the influenza virus hemagglutinin protein. MultiBac, an advanced baculovirus system, has been widely adopted in the last decade to produce multiprotein complexes with many subunits that were hitherto inaccessible, for academic and industrial research and development (Sari et al., 2016). Baculoviruses, modified to contain mammalian promoters (BacMam viruses), have proven to be efficient gene delivery vectors for mammalian cells and provide an alternative transient mammalian cell based protein expression approach to that of plasmid DNA based transfection methodologies (Kost and Kemp, 2016).
One of the most appealing features of baculovirus–insect expression systems has been the eukaryotic protein processing capabilities of the host. Accordingly, these systems are widely considered to be excellent tools for recombinant glycoprotein production. Nearly a thousand of high-value foreign proteins have been successfully produced in the system and the insect baculovirus may be applied in production of vaccines (Kang, 1997), gene therapy (Ghosh et al., 2002) and recombinant baculovirus insecticides (Assenga et al., 2006).
Like any other system, this system is also not free from limitations. As the host cell infected with nuclear polyhedrosis virus will eventually die, the heterologous gene cannot be expressed continuously. Every round of synthesis of the protein of interest requires the infection of new insect cells. Therefore, this system is inferior to prokaryotic and yeast systems in terms of its capacity for continuous fermentation. Moreover, insect cells and mammalian cells differ in their glycosylation patterns, such as in the lengths of oligosaccharides and in mannose content (Kost and Condreay, 1999; Marheineke et al., 1998), so the bioactivity and immunogenicity of insect expression products are somewhat different from those of the natural product. However, people have successfully addressed this limitation by genetically transforming established lepidopteran insect cell lines with constitutively expressible mammalian genes. This approach has yielded transgenic insect cell lines with normal growth properties that can support baculovirus infection, having new N glycan processing enzyme activities, producing humanized recombinant glycoproteins (Donald, 2003).
Mammalian Systems
There appears to be a progressive increase in the application of mammalian cells for protein production. Expression systems utilizing mammalian cells for recombinant proteins are able to introduce proper protein folding, post-translational modifications and product assembly, which are important for complete biological activity (Khan, 2013). They also promote signal synthesis, process and can secrete and glycosylate proteins, particularly eukaryotic proteins.
A number of mammalian cell lines have been utilized for protein expression with the most common being HEK 293 (Human embryonic kidney) and CHO (Chinese hamster ovary). These cell lines can be transfected using polyethyleneimine (PEI) or calcium phosphate. Apart from this, baby hamster kidney (BHK) cells, Vero cells, mouse L-cells and myeloma cell lines like J558L and Sp2/0, etc., are also employed as hosts for the establishment of stable transfectants (Geisse et al., 1996; Castro et al., 2014). Even though, the high cost, complicated technology and potential contamination with animal viruses have been the limitations for its use in large-scale industrial production, this system is often utilized to express many heterologous proteins including viral structural protein and bioactive peptide for specific functional analysis because of its advantages (Nagpal et al., 2004). Mammalian systems such as CHO and BHK cell systems are the ideal choice for production of therapeutic proteins as these are capable of glycosylating the protein at the correct sites. However, cost of production of the products using these cell systems is high because of the slow growth and expensive nutrient requirement. The choice of an expression system invariably influences the character, quantity and cost of a final product.
Further, recent advances have been made in producing therapeutic proteins by using transgenic animates. Eukaryotic individuals systems are a newly emerging expression system, which includes both individual animal and individual plant expression systems.
Transgenic Plants
The plants can be considered as a solar- powered bioreactor and proved to be advantageous over the alternative fermentation systems of biomass production using microbial or animal cells. The requirements for plant system are rather simple and inexpensive. Plants, being eukaryotic, are also capable of the post-translational processing of proteins of eukaryotic origin, which may be essential to their proper functioning. The complex, multi-meric proteins can be readily assembled in plant cells and individual plant expressing genes encoding different components of multi-meric complexes can be readily obtained by sexual crossing of plants harbouring a single transgene. The use of genetically engineered plants to produce valuable proteins is increasing slowly. The system has potential advantages of economy and scalability. However, variations product yield, contamination with agrochemical and fertilizers, impact of pest and disease and variable cultivation conditions should also be considered. Plant cell culture system combines the advantages of whole plant system as well as animal cell culture, Although no recombinant products have yet been produced commercially using plant cell culture several companies are investigating the commercial feasibility of such a production system (Balamurugan et al., 2006).
Plant cells share some architectural and functional similarities with animal cells. In particular, they constitute an optimal system to express heterologous proteins that require complicated post-translational modifications, such as some glycoproteins, bioactive peptides, and drugs. Unlike most other expression systems, individual plant expression systems have greater and distinctive flexibility and utility. Firstly, the heterologous proteins expressed can be localized to different organs of the plant by controlling the tissue-specific regulatory sequences involved in gene expression. Secondly, proteins can be expressed at specific growth stages by manipulating the inducible components and development-specific regulatory sequences. Thirdly, plants can be grown in the field, providing a very inexpensive source of material compared to any organism that needs to be grown in fermentors. Thus scale-up of plant-based expression is much easier than in other systems: more plants can be grown easily, reliably increasing total yield, compared to culture and fermentor reactor-based systems that are often very difficult to scale up (Chen and Davis, 2016). Finally, expression of proteins in plant seeds results in a unit of production (seed), in which proteins are extremely stable, readily stored, and easily extracted and purified (Yin et al., 2007; Yemets et al., 2014).
Table 1: Merits and demerits of different host systems for expression of recombinant proteins
| Host system | Merits | Demerits |
|
Escherichia coli |
Easy Quick Economical Rapid growth rate Capacity for continuous fermentation |
Does not possess necessary machinery for removing introns from transcripts Foreign gene might contain sequences that act as termination signals resulting in premature termination and loss of gene expression Codon bias Lack of post translational modifications Glycosylation is extremely uncommon in bacteria Production of proteins in the insoluble form or in the form of inclusion bodies Degradation of proteins Accumulation of endotoxins |
|
Bacillus subtilis |
Does not produce LPS/endotoxins Can be transformed readily with many bacteriophages and plasmids Capable of secreting functional extracellular proteins directly into the culture medium |
Production of extracellular proteases which can recognize and degrade heterologous proteins Instability of plasmids Reduced or non expression of the protein of interest |
|
Yeast system |
Rapid growth in low cost medium Appropriate post-translational modifications Safety of the system is guaranteed No endotoxins production |
Hyperglycosylation of proteins Codon bias Inefficient in secreting the proteins into growth medium leading to intracellular retention |
| Filamentous fungus |
High-level of expression |
Complex Lack of knowledge on physiology |
| Baculovirus /Insect system |
High level of expression Appropriate posttranslational modifications Safe for vertebrates Excellent tool for recombinant glycoprotein production |
Continuous expression not possible More demanding culture conditions |
| Mammalian cells / system |
Proper protein folding Appropriate post-translational modifications and product assembly Proper glycosylation |
High cost Complicated technology Potential contamination with animal viruses |
| Transgenic plants |
Easy scaling up at low cost Proteins can be localized to different organs at different growth stages High yield |
Expression levels are target dependent Functional assays yet to be developed |
| Transgenic animals |
Proper protein folding Appropriate post-translational modifications and product assembly Proper glycosylation |
Relatively longer production period Low yield Higher costs |
Transgenic Animals
Transgenic animal bioreactors can produce therapeutic proteins with high value for pharmaceutical use. The mammary gland has generally been considered the organ of choice to express valuable recombinant proteins because milk is easily collected in large volumes and is the best available bioreactor. Foreign proteins are commonly reported to be produced in transgenic milk at rates of several grams per litter. Apart from milk, egg white, blood, urine, seminal plasma and silkworm cocoon are the alternative systems available for the production of useful pharmaceutical proteins. The various mammals used as bioreactors are rabbits, pigs, sheep, goats and cows. Each of these species offers advantages and drawbacks. Rabbits are sufficient to produce several kilograms of proteins per year. The rabbit is particularly flexible, allowing rapid generation and scaling-up. For very high protein production, larger animals are needed
(Wang et al., 2013). However, the individual animal expression system requires a relatively longer production period with low yield and higher costs than above-mentioned expression systems. So this system can express foreign proteins mainly for medical purposes (Yin et al., 2007).
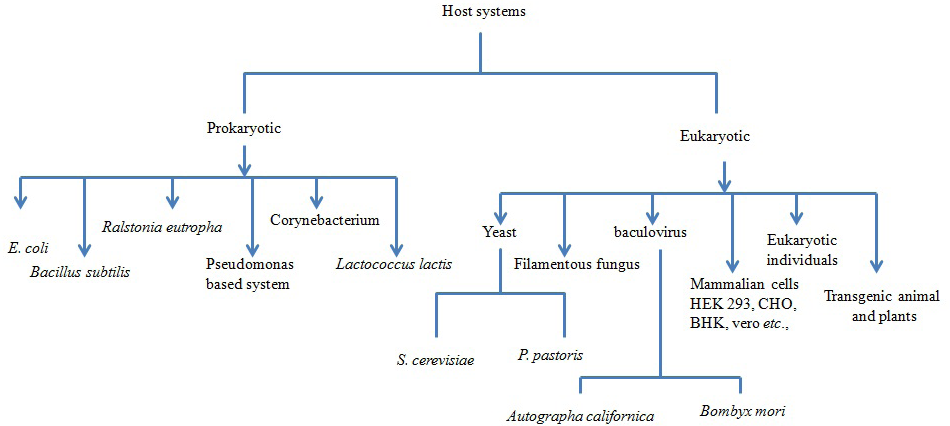
In nutshell, the different host systems available for expression of recombinant proteins and their merits and demerits are summarized (Figure 1 and Table 1). Recombinant proteins are being produced using either of the heterologous systems mentioned above and have a potential value in the development of diagnostic test as well as vaccines for the prophylaxis of various infectious diseases of human and veterinary importance (Balamurugan et al., 2006). A hallmark in the prevention and control of several diseases of the animals and human used by viruses, bacteria and parasites is the development of suitable vaccines against infectious diseases. So far, various attempts have been made in the recent past to produce antigens in heterologous systems for use as diagnostics as well as prophylactics. Biologically active peptides and proteins have many potential applications including being used as vaccines, immuno-modulators, growth factors, hormones and enzymes.
Conclusions
In general, rDNA technology has indeed made tremendous breakthrough in the discovery of various recombinants antigens or proteins. The new generation proteins prepared from the viral/microbial proteins; their fragments or the nucleic acid sequences have been attractive because of their stability, non-infectious nature, homogeneity as well as their cost-effectiveness. One should carefully choose the system for a specific expression procedure considering bio-characteristics of the protein, quality and quantity of the protein, cost, availability, convenience and purposes of the expressed products. Further, for functional analysis and preparation of vaccines eukaryotic system can be considered and for production of artificial antigens for diagnostic purposes or for studying the structural analysis of particular proteins that require no post-translational modification, prokaryotic system can be selected. Products developed in the field of veterinary medicine will be most
valuable for further development of rDNA products in the coming decades. In view of high market potential for recombinant therapeutics as the case with human therapeutics, indigenous technology should also be developed for the veterinary field to develop a prophylactics and diagnostics. This can be achieved by strengthening the linkages among various institutes having expertise in different disciplines related to rDNA technology and increased interaction with the industry.
Acknowledgements
All the authors who contributed to the subject irrespective of their citation being figured or not are acknowledged.
Conflict of Interest
No conflicts of interests are declared by authors for the contents in this manuscript.
Authors’ contribution
A. R. Gomes drafted the manuscript, S. M. Byregowda and B. M. Veeregowda gave the technical guidance. V. Balamurugan provided technical guidance, inputs and edited the manuscript.
References