Advances in Animal and Veterinary Sciences
Case Report
Histopathology of the Organs from Cattle with Leptospirosis
Yos Adi Prakoso1*, Sitarina Widyarini2, Kurniasih Kurniasih2
1Faculty of Veterinary Medicine, University of Wijaya Kusuma Surabaya, East Java, Indonesia, 60225; 2Department of Pathology, Faculty of Veterinary Medicine, University of Gadjah Mada, Yogyakarta, Indonesia, 55281.
Abstract | Leptospirosis is one of the neglected diseases distributed worldwide. Leptospirosis causes severe infection in cattle. A total of four male cattle were found dead in the cowshed during July 2019 – April 2020. The necropsy procedure was conducted, and various macroscopic lesions were identified. Further, several fresh and fixed specimens were collected and transported to the laboratory for ELISA and histopathology. The qualitative ELISA showed that all kidney samples were positive against Leptospira IgG. The histopathology showed that the lung specimens were suffering from pneumonia (3/4), the liver congestion (2/4), liver fatty degeneration (1/4), and chronic nephritis and glomerulonephritis (4/4). Based on the Gram staining, all kidney samples showed the appearance of slender form bacteria that may belong to Leptospira sp in the lumen of tubules. All dead cattle showed moderate and minimal expression of CD4+ and CD8+ in the lung and liver concomitantly. In contrast, there is no expression of CD4+, and a high expression of CD8+ was observed from the kidneys. This study demonstrated that leptospirosis in cattle impacts several organs with severe manifestation occurs on the kidneys.
Keywords | Cattle, ELISA, Histopathology, Immunohistochemistry, Leptospirosis.
Received | October 07, 2019; Accepted | February 17, 2020; Published | September 05, 2020
*Correspondence | Yos Adi Prakoso, Faculty of Veterinary Medicine, University of Wijaya Kusuma Surabaya, East Java, Indonesia, 60225; Email: yos.vet.docter@gmail.com
Citation | Prakoso YA, Widyarini S, Kurniasih K (2020). Histopathology of the organs from cattle with leptospirosis. Adv. Anim. Vet. Sci. 8(11): 1220-1224.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.11.1220.1224
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Prakoso et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Leptospirosis is one of the neglected diseases distributed worldwide. Leptospirosis is caused by the bacteria from a genus of Leptospira sp. Leptospira sp. has been reported as a zoonotic bacteria that can be transmitted from animal to human or vice versa. Moreover, leptospirosis does not only occur in the rural but also in the urban area. It because rodents act as the primary reservoir of this disease (Boey et al., 2019). Rodent spread the bacteria through the urine to the environment. The presence of the bacteria in the environment promotes direct and indirect exposure that leads to an infection to humans with various clinical manifestations (Goarant, 2016).
Leptospirosis can cause systemic infection as acute and chronic types in cattle (Zarantonelli et al., 2018). The pathogenic impact of leptospirosis in cattle is severe repro
ductive disorders, abortion (Favero et al., 2017), stillbirth, and weak off-spring (Suwancharoen et al., 2016). Further, it promotes the economic losses in the livestock industry. In this case, we reported the untypical histopathology and immunohistochemistry of several organs from the cattle with leptospirosis.
Case presentation
Anamnesis and Necropsy
Four male cattle were found dead in the local cowshed, sub-district Krembung, East Java, on July 2019 – April 2020. Further, the disinfection was conducted in the cowshed area, and the necropsy was performed. Various gross lesions were found from the dead cattle. The details of the case history of the dead cattle were embedded in Table 1.
Table 1: Case history of dead cattle with leptospirosis
| Case code | Breed | Sex, age | Obit | Clinical signs | Gross lesions |
| C1 | Bali cattle | M, 2 y | 18 July 2019 |
Anorexia Hematuria |
Redness in lung Adhesion of renal capsule |
| C2 | Limousine | M, 3 y | 26 July 2019 | Hematuria |
Redness in lung Liver enlargement Paleness of kidney |
| C3 | Bali cattle | M, 2 y | 8 August 2019 | No clinical signs | Paleness of kidney |
| C4 | Cross breed | M, 2 y 5 m | 1 April 2020 | No clinical signs |
Redness in lung Adhesion of renal capsule Asymmetric size of kidney |
M = male, y = year, m = month.
Table 2: Histopathology scoring system
| Score | H&E | Gram and IHC | |||
| Severity | Duration | Distribution | Exudate | ||
| 0 | NHC | NHC | NHC | NHC | NI |
| 1 | Minimal | Acute | Focal | Suppurative | Minimal |
| 2 | Mild | Chronic | Multifocal | Fibrinous | Mild |
| 3 | Moderate | Chronic active | Locally extensive | Necrotizing | Moderate |
| 4 | Severe | - | Diffuse | Fibrinopurulent | Severe |
| 5 | - | - | - | Granulomatous | - |
NHC = no histopathological changes, NI = not identified, - = not categorized.
Samples Collection
The lung, liver, and kidney were collected and separated into two parts. The first part was stored inside the sterile plastic bag and cooled at 4° C. The second part was stored using 10% neutral buffer formalin (NBF). The specimens were transported to the Integrated Laboratory, University of Muhammadiyah Sidoarjo, East Java, Indonesia.
Enzyme-linked Immunosorbent Assays (ELISA)
The fresh specimens were tested using ELISA against Leptospira IgG (LEBTOSPIRA IgG ELISA kit, Cat. No: MBS036135, MyBioSource). Before the ELISA, the organs were mashed up using the mortar and mixed with sterile distilled water. The mixture was then centrifuged at 3000 rpm for 5 minutes, and it repeated three times. The ELISA was performed following the provided procedure by the manufacturer. The optical density (OD) was read using an ELISA reader at 450 nm of wavelength. The samples were categorized as positive (OD ≥ 1.1) and negative (OD ≤ 1.0).
Histopathology and Immunohistochemistry
The fixed specimens were processed for histopathology using several staining, including hematoxylin (Harris Hematoxylin Nuclear Stain, Cat. No: 3801560, Leica) and eosin (Eosin Secondary Counter Stains, Cat. No: 3801600, Leica) staining, Gram (Remel Gram stain kit, Cat. No: R40080, Thermo Fisher Scientific), and immunohistochemistry against CD4+ (RTU-CD4-1F6, Cat. No: PA0427, Leica), and CD8+ (RTU-CD8-295, Cat. No: PA0183, Leica). The H&E was used as routine staining, Gram used to stain bacteria, and IHC used to express the immunoreactivity of CD4+ and CD8+. The H&E and IHC staining were conducted following the procedure provided by manufacturer. However, the Gram staining was performed following the previous study (Becerra et al., 2016).
Morphometry
The H&E slide was analyzed by a single pathologist using the scoring system. The score was calculated based on the addition of scores from several parameters include severity, duration, distribution, and types of exudation. The Gram and IHC staining slide was scored based on the distribution of the staining target (Table 2).
Results and Discussion
The result showed that all the kidney specimens were positive against Leptospira IgG (Table 3). The kidney samples were tested against Leptospira IgG because these are the predilection of the Leptospira sp. The appearance of IgG from the kidneys indicated that all cattle infected by Leptospira sp. Further laboratory tests revealed various histopathological changes of the lung, liver, and kidney.
Table 3: ELISA of Leptospira IgG of kidney from dead cattle with leptospirosis
| Organ | Case code | |||
| C1 | C2 | C3 | C4 | |
| Kidney | + | + | + | + |
Table 4: Histopathology of lung, liver and kidney from dead cattle with leptospirosis
| Organ | Parameter | Case code | |||
| C1 | C2 | C3 | C4 | ||
| Lung | H&E score | 10 | 3 | 2 | 13 |
| Gram score | 0 | 0 | 0 | 0 | |
| CD4+ score | 2 | 1 | 3 | 2 | |
| CD8+ score | 1 | 1 | 2 | 4 | |
| Morphologic diagnosis | Severe chronic diffuse pneumonia | Minimal acute focal pneumonia | Pulmonary congestion | Severe chronic active diffuse fibrinous pleuro-pneumonia | |
|
Liver |
H&E score | 0 | 4 | 0 | 3 |
| Gram score | 0 | 0 | 0 | 0 | |
| CD4+ score | 0 | 0 | 1 |
0 |
|
| CD8+ score | 0 | 2 | 0 | 0 | |
| Morphologic diagnosis | NHC | Congestion, Fatty liver degeneration | NHC | Liver congestion | |
| Kidney | H&E score | 10 | 7 | 5 | 14 |
| Gram score | 4 | 2 | 3 | 4 | |
| CD4+ score | 0 | 0 | 0 | 0 | |
| CD8+ score | 4 | 4 | 4 | 3 | |
| Morphologic diagnosis | Severe chronic diffuse interstitial nephritis | Moderate chronic multifocal glomerulo-nephritis | Mild chronic focal interstitial nephritis | Severe chronic active diffuse necrotizing glomerulo-nephritis | |
NT = not tested, + = positive, - = negative, NHC = no histopathological changes.
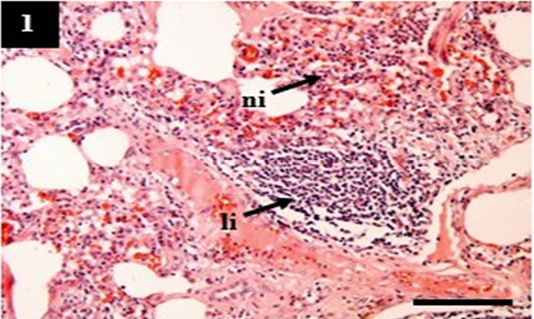
Figure 1: Histopathology of lung from case C1 showed predominantly infiltration of neutrophil (ni) and lymphocytes (li) within the pulmonary interstitial. H&E, 10×.
The H&E score showed the severity of histopathology. A higher score indicated increasingly severe histopathological changes (Table 4). The lung showed pneumonia with various manifestations that belong to acute (C2), chronic (C1), and chronic active (C4). It was suspected that tissue destruction of the lung in C1, C2, and C4 reflected grad
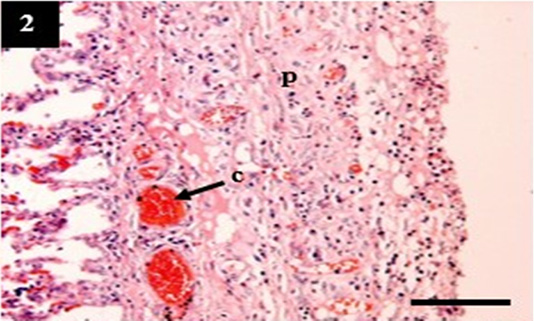
Figure 2: Histopathology of lung from case C4 showed the thickening of the pleural wall (p) and congestion within the venous (c). H&E, 10×.
ual pathogenesis in leptospirosis (Figures 1 and 2). However, the liver did not show significant histopathological changes (Figure 3). Further, the kidney from C1 and C3 showed chronic interstitial nephritis with diffuse and focal distribution, respectively. Besides, the C2 and C4 showed chronic glomerulonephritis (Table 4, Figure 4, and 5).
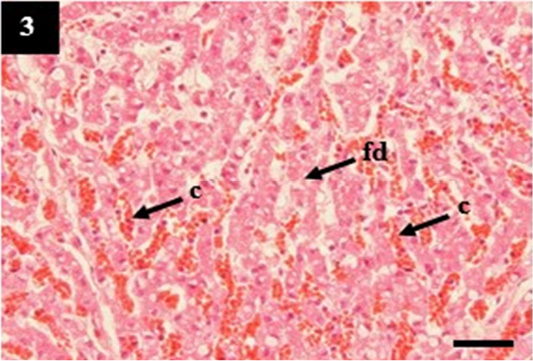
Figure 3: Histopathology of liver from case C2 showed sinusoidal congestion (c) with a moderate fatty degeneration (fd) in the cytoplasm of hepatocytes. H&E, 40×.
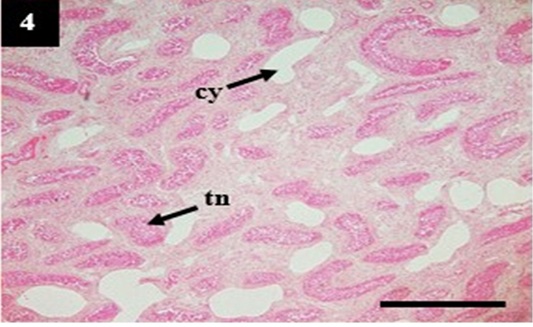
Figure 4: Histopathology of kidney from case C4 showed moderate cystic dilation of tubules (cy) and tubular necrosis (tn). H&E, 4×.
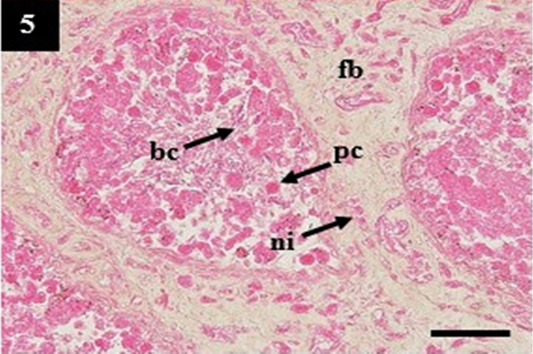
Figure 5: Histopathology of kidney from case C4 showed the deposit of pinkish homogenous (pc) within the lumen that may belong to amyloid, the thickening of fibrous tissue (fb) and infiltration of neutrophil (ni) within the interstitial. H&E, 20×.
Gram staining of lung and liver did not show the presence of slender form bacteria that may belong to Leptospira sp. In contrast, the kidney showed a high presentation of slender form bacteria within the lumen (Table 4 and Figure 6). Surprisingly, it is synergistic to the immunohistochemistry score. The zero scores of Gram staining in the lung showed the appearance of CD4+ and CD8+ as normal lymphocytes within the nodular lymphoid tissue. It was different from the kidney samples. The kidney showed a high expression of CD8+ as the compensatory impacts of Leptospira sp infection (Table 4).
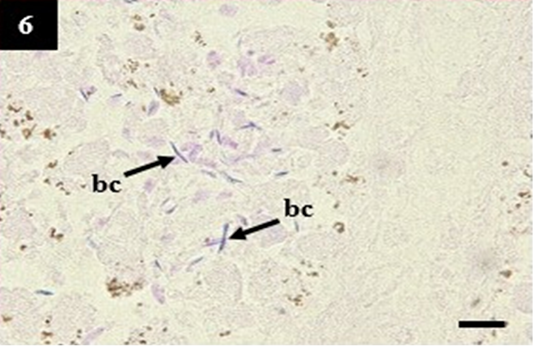
Figure 6: Histopathology of kidney from case C1 showed the slender form bacteria in the lumen that may belong to Leptospira sp. (bc). Gram, 1000×.
Leptospira sp. is a pathogenic bacteria that can survive in the environment for prolonged periods. The ability of Leptospira sp. to survive in the environment enables this bacteria to cause direct and indirect infection in humans and animals. Leptospirosis commonly occurs and spreads in the unsanitary area with a high population of rats. Wild rats act as the main reservoirs of Leptospira sp. that promote freshwater contamination through its urine (Pellizzaro et al., 2019). Furthermore, it enables the waterborne disease in cattle.
Leptospira sp. is utilizing its virulence factors such as surface protein TlyC to bind the extracellular matrix of host tissue and erythrocytes (Evangelista and Coburn, 2010). In advances, this bacteria circulates through the bloodstream and releases sphingomyelinase C gene A (sphA) that induces endothelial and membrane cell damage (Narayanavari et al., 2015). In a healthy immunological response, the host body releases T-lymphocytes (CD4+ and CD8+) to eliminate the circulatory antigen. However, both CD4+ and CD8+ are suppressed during leptospirosis because leptospirosis can depress innate immune response associated with a decrease in NK cell, and adaptive immune response associated with an inhibition of CD4+ and CD8+ lymphocytes (Duarte-Neto et al., 2019).
The synergic mechanism between Leptospira virulence factors and its ability to inhibit immune response generates severe multisystem infection, including hepatic dysfunction, pulmonary lesion, and renal failure (De Brito et al., 2018). All those histopathological changes are similar to this study. The chronic pathogenesis of cattle leptospirosis in this study may be due to the presence of circulatory IgG. However, the IgG protection status in this study cannot be concluded because it only used qualitative ELISA.
Significantly, this study demonstrated that modified tissue Gram staining could be used as a diagnostic tool to stain the presence of slender form bacteria that may belong to Leptospira sp. In this study, Leptospira sp. stained purple that similar to the Gram-positive bacteria because it has a surface membrane that shares a feature of Gram-positive and Gram-negative bacteria such as the murein cell wall and lipopolysaccharides, respectively (Ko et al., 2009). Unfortunately, this study only used qualitative ELISA as a comparison against Gram staining. Further study is necessary to be performed to compare the sensitivity and specificity of Gram staining between the existence of Leptospira sp. within the tissue using a more considerable number of specimens and a more sophisticated laboratory test.
Conclusions
This study demonstrated that leptospirosis in cattle impacts on the lung, liver, and kidney with various manifestations. The significance of this study is that we found the modified Gram staining could be used to stain the slender form bacteria that may belong to Leptospira sp. even though further exploration must be conducted.
Acknowledgment
All staff from Integrated Laboratory, UMSIDA, East Java, Indonesia were acknowledged for their assistance during the processing of the collected specimens.
Author’s contribution
All the authors contributed equally during this study and approved the final version of the manuscript.
Conflict of interests
The author declares that there is no conflict of interest.
References






