Advances in Animal and Veterinary Sciences
Research Article
Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Assay for the Detection of Rabies Virus
Rachamreddy Venkata Chandrasekhar Reddy1,2, Alapati Krishna Satya2, Kota Sri Naga Leela Surendra1, Samir Kumar Rana1, Bhaskaran Mohana Subramanian3, Girish Kumar Sharma4, Villuppanoor Alwar Srinivasan5*
1National Dairy Development Board, R and D Laboratory, Hyderabad, Telangana – 500032, India; 2Department of Biotechnology, Acharya Nagarjuna University, Guntur, Andhra Pradesh - 522004, India; 3Research and Development Center, Biological E limited, Hyderabad, Telangana – 500032, India; 4National Dairy Development Board, Anand, Gujarat- 388001, India; 5National Dairy Development Board, 33-Telecom Nagar, Gachibowli, Hyderabad, Telangana – 500032, India.
Abstract | A one-step real-time reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay was developed for detecting rabies virus (RABV) genome. RT-LAMP primers were designed based on the most conserved region of N gene sequences covering various rabies virus isolates of India and representative sequences from each genetic lineage of rabies virus around the world. Degenerate bases were included in the RT-LAMP primers depending on the sequence variation among the isolates. Amplification products of the RT-LAMP assay was monitored by real-time turbidimeter, agarose gel electrophoresis and visual colour change of HNB dye. Detection limit of the RT-LAMP assay was 103 RNA copies or 10-1 FFU (0.1 Fluorescence focus units) of rabies virus particles per reaction. A total of 45 known rabies positive clinical tissues were tested using the RT-LAMP assay and all these samples were confirmed as rabies by all the three detection methods of RT-LAMP assay. Both diagnostic specificity and sensitivity of the assay were 100% comparable to RT-PCR. Thus this technique is an excellent tool for diagnosis and epidemiological studies of rabies, where the disease is enzootic which can be carried out with minimum laboratory facilities.
Keywords | Rabies virus, RT-LAMP, HNB dye, RT-PCR, India
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | October 27, 2016; Accepted | November 01, 2016; Published | November 08, 2016
*Correspondence | Dr. Villuppanoor Alwar Srinivasan, Advisor (Animal Health), National Dairy Development Board, 33-Telecom Nagar, Gachibowli, Hyderabad-500032, India; Email: srinivasanva1948@gmail.com
Citation | Reddy RVC, Satya AK, Surendra KSNL, Rana SK, Subramanian BM, Sharma GK, Srinivasan VA (2016). Reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of rabies virus. Adv. Anim. Vet. Sci. 4(11): 584-592.
DOI | http://dx.doi.org/10.14737/journal.aavs/2016/4.11.584.592
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Reddy et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Rabies is one of the zoonotic viral diseases with public health significance and causes fatal encephalitis. Although the disease can be caused by all members of the genus Lyssavirus, the majority of cases reported are due to the prototype species, classical rabies virus (RABV) (ICTV, 2009). Rabies virus is endemic in 150 countries, with the exception of few countries which include New Zealand, Australia, the United Kingdom and Japan. Most of the developed countries have succeeded in controlling rabies in domestic dogs by implementing the appropriate control measures. While, rabies in stray dogs is a major concern for public health in many developing countries (WHO, 2005) sylvatic rabies is commonly encountered in developed countries.
Early identification of the virus in a suspected rabid animal is crucial for the appropriate advocation of post-exposure prophylaxis in humans and also essential for the effective control of the disease. Currently, several rabies detection techniques have been developed, which includes fluorescent antibody test (FAT), virus isolation in cell culture or mouse inoculation test (MIT), RT-PCR and real-time PCR assays (WHO, 2005; Dean et al., 1996; Heaton et al., 1997; Nadin-Davis, 1998; Wakeley et al., 2005; Nagaraj et al., 2006). However, all the techniques are time consuming and require sophisticated laboratory facilities.
Invention of loop mediated isothermal amplification (LAMP) provides an alternative for molecular diagnosis. Using this approach, amplification of nucleic acids under isothermal conditions (e.g., water bath) is rapid with high specificity and efficiency (Notomi et al., 2000). One of the major advantages of this technique is the simplicity of the test. Application of the LAMP assay for clinical diagnosis of infectious diseases was previously established (Notomi et al., 2000; Enosawa et al., 2003; Poon et al., 2005; Shanti et al., 2007; Madhanmohan et al., 2013). Subsequent to that rabies diagnosis based on the RT-LAMP assay has also been described and evaluated earlier in the Philippines, Brazil, Ghana and Zambia (Boldbaatar et al., 2009; Saitou et al., 2010; Hayman et al., 2011; Muleya et al., 2011).
The amplification efficiency of the RT-LAMP method is extremely high due to continuous amplification under isothermal conditions which results in the production of a large amount of target DNA/RNA as well as a large amount of the by-product magnesium pyrophosphate, which leads to turbidity (Mori et al., 2001). Therefore, quantitative detection of gene amplification is possible by real-time monitoring of the turbidity in an inexpensive turbidi-meter. In addition, the higher amplification efficiency of the RT-LAMP method enables simple visualization with the naked eye by adding metal ion indicator dye (HNB) or intercalating dyes, such as SYBR Green I (Parida et al., 2005; Goto et al., 2008). Therefore, Reverse transcription Loop mediated Isothermal Amplification (RT-LAMP) method has emerged as a powerful technique for the detection of microbial infections and the method is widely accepted as a diagnostic tool in the field of veterinary and medical sciences (Fu et al., 2011).
In the present study, an attempt was made to develop RT-LAMP assay for the detection of rabies virus strains belonging to different lineages circulating globally. Subsequently, we evaluated this tool for the detection of rabies virus directly from clinical samples collected from different geographical regions in India as well as with fixed strains. The results were compared with one step RT-PCR assay.
Materials and Methods
Cell Line and Laboratory Strains of Rabies Virus
Mouse neuroblastoma (MNA) cells maintained in the laboratory were used in this study. Two laboratory (fixed)
Table 1: Rabies isolates used in this study and the results of RT-LAMP and One step RT-PCR
|
Virus ID |
Host |
Genetic lineage |
RT-LAMP |
One step RT-PCR |
|
IAP-R91 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IKE-R73 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IKE-R77 |
Cow |
Arctic-like |
+Ve |
+Ve |
|
IKE-R78 |
Cow |
Arctic-like |
+Ve |
+Ve |
|
IKE-R86 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R87 |
Calf |
Arctic-like |
+Ve |
+Ve |
|
IMA-R88 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R94 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R97 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R101 |
Cattle |
Arctic-like |
+Ve |
+Ve |
|
IKE-R106 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R107 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R109 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R110 |
Cattle |
Arctic-like |
+Ve |
+Ve |
|
IKE-R111 |
Goat |
Arctic-like |
+Ve |
+Ve |
|
IKE-R114 |
Cow |
Arctic-like |
+Ve |
+Ve |
|
IKE-R116 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKE-R121 |
Goat |
Arctic-like |
+Ve |
+Ve |
|
IKA-R129 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKA-R132 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKA-R142 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IKA-R144 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IMA-R146 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
ITN-R148 |
Dog |
Sub-continent |
+Ve |
+Ve |
|
IKE-R154 |
Elephant |
Sub-continent |
+Ve |
+Ve |
|
IKE-R155 |
Cow |
Sub-continent |
+Ve |
+Ve |
|
IMA-R189 |
Human |
Arctic-like |
+Ve |
+Ve |
|
IAP-R190 |
dog |
Arctic-like |
+Ve |
+Ve |
|
IAP-R191 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IAP-R192 |
Dog |
Arctic-like |
+Ve |
+Ve |
|
IAP-R193 |
Human |
Arctic-like |
+Ve |
+Ve |
|
IAP-R194 |
Human |
Arctic-like |
+Ve |
+Ve |
|
IAP-R195 |
Human |
Arctic-like |
+Ve |
+Ve |
|
IAP-R196 |
dog |
Arctic-like |
+Ve |
+Ve |
|
IUP-R197 |
horse |
Arctic-like |
+Ve |
+Ve |
|
IUP-R198 |
horse |
Arctic-like |
+Ve |
+Ve |
|
IGR-R199 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IGR-R200 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IGR-R201 |
Nilgai |
Arctic-like |
+Ve |
+Ve |
|
IGR-R202 |
Mongoose |
Arctic-like |
+Ve |
+Ve |
|
IGR-R203 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IGR-R204 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IGR-R205 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IGR-R206 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
IGR-R207 |
Buffalo |
Arctic-like |
+Ve |
+Ve |
|
PV |
Vaccine strain |
Cosmopolitan |
+Ve |
+Ve |
|
CVS |
Vaccine strain |
Cosmopolitan |
+Ve |
+Ve |
strains of rabies virus (challenge virus standared-CVS-11 and Pasteur virus strain-PV) were included in the present study for standardization of the assay. The viral strains were propagated in MNA cells and titers of the virus were determined as described earlier by Abelseth and Atanasiu (1996).
Clinical Samples
Rabies virus (RABV) isolates used in the present study were derived from clinical brain tissues from animals or humans. A total of 45 samples (Arctic-like 1 lineage: n=42 and sub-continental lineage: n= 3) originated from nine different species of terrestrial mammals (Table 1) were used. All these samples were tested positive for the presence of RABV-specific antigen and rabies virus genome by FAT and one step RT-PCR respectively (Dean et al., 1996; Nagarajan et al., 2009). These known positive clinical samples maintained in the repository of this laboratory were used in the current study. Normal Swiss mouse brain samples (n = 10) were also included as negative control to rule out the possibility of false positive reactions.
RNA Extraction
Total RNA was extracted from brain tissue homogenate obtained from clinical samples, as well as CVS and PV viral strains propagated in MNA cells using TriZol reagent following the manufacturer’s protocol (Invitrogen, USA). Subsequently, RNA samples were stored at -80°C until further use.
RT-LAMP Primer Design
RT-LAMP primers used in the study were designed using LAMP designer 1.10 software (http://www.premierbiosoft.com/isothermal/lamp.html), based on the nucleoprotein gene (N) sequence of the rabies virus (GenBank Accession Number: M13215). A set of six primers comprising two outer, two inner and two loop primers that recognize eight distinct regions on the target sequence was designed (Notomi et al., 2000; Nagamine et al., 2002).
To design the RT-LAMP primers, N gene sequence of RABV belonging to different lineages around the world was retrieved from GenBank and multiple sequence alignment was performed using MEGA 6.0 software (Nagarajan et al., 2009; Reddy et al., 2015). Though, the N gene is one of the conserved region in rabies virus, our sequence analysis of all the lineages have found few mismatches. The RABV N gene sequences included to design degenerated primers are Arctic-like 1 lineage (KX434483-KX434505, KX434509-KX434523, EF437215, AB699220, EU086162, EU086198, JX944570, JX944565), Arctic-related lineage (U22655, U22656, AY730597, AY730596, GU937039); Sub-continental lineage (KX434506- KX434508, AB041968, AB041969, AB041965), Cosmopolitan lineage (Vaccine strains; CVS-11 and PV), African (U22628, U22640) and Asian lineage (EU086176, EU086177, EU086190, EU086192). The RT-LAMP primers were modified by incorporating degenerate bases wherever necessary. These primers are labeled as Deg-RT-LAMP primers and used in the present RT-LAMP assay. The details of the Deg-RT-LAMP and RT-PCR primers are in Table 2, and a schematic representation of the RT-LAMP assay primer design is also shown in Figure 1.
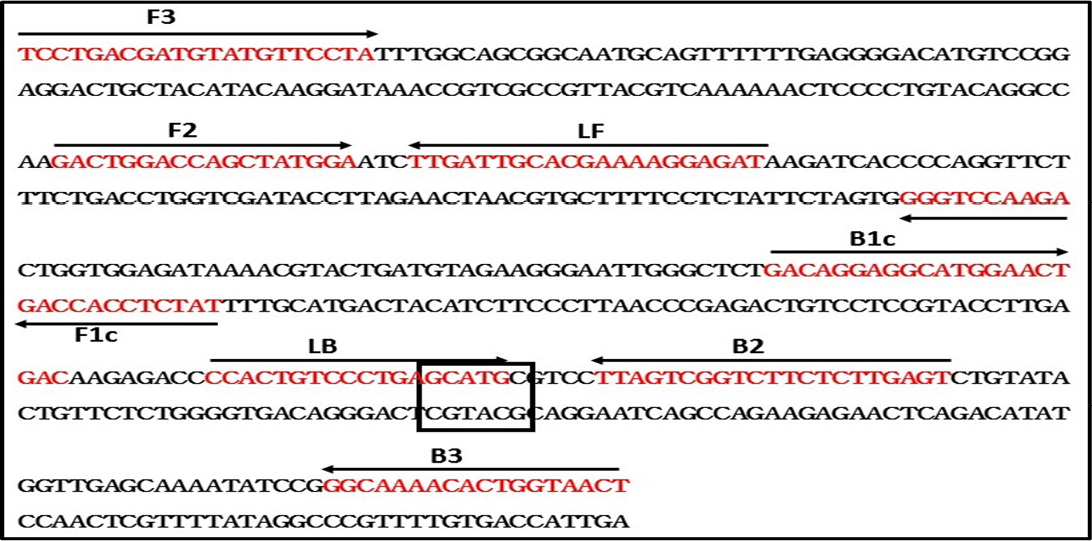
Figure 1: Schematic diagram of primers used in the RT-LAMP assay and the nucleotide sequence of the Nucleoprotein gene used to generate the inner, outer and loop primers (GenBank accession No. M13215). The arrows indicate the location of primers and the direction of primer extension. Sph1 restriction enzyme site was highlighted
Table 2: Details of the primers used for RT-LAMP study
|
Method |
Primer name |
Genome position |
Sequence (5′ - 3′) |
Length (bp) |
|
Deg-RT-LAMP |
Deg-F3 |
268-288 |
YCCWGATGATGTRTGYTCCTA |
21 |
|
Deg-B3 |
534-551 |
AGTTRCCRGTGTTYTGYC |
18 |
|
|
Deg-FIP (F1c+F2) |
382-403+332-349 |
TATYTCYACMAGAGAATCYGGR+GAYTGGACCAGCTAYGGR |
40 |
|
|
Deg-BIP (B1c+B2) |
436-456+488-508 |
GACNGGAGGAATGGARYTRAC+ACTCAARAGAAGACRACTAA |
42 |
|
|
Deg-LF |
353-373 |
RTCYCCTTTYCKTGCRATCAR |
21 |
|
|
Deg-LB |
465-482 |
CCACTGTYYCYGAGCATG |
18 |
|
|
One Step RT- PCR |
N-FP |
55-73 |
ATGTAACACCTCTACAATG |
19 |
|
N-RP |
641-660 |
CAATTAGCACACATTTTGTG |
20 |
RT-LAMP Assay
The RT-LAMP assay was carried out in a total of 25-µl reaction volume using a Loopamp RNA Amplification Kit (Eiken Chemical, Tokyo, Japan), containing 5 pmol each of outer primers F3 and B3, 40 pmol of internal primers FIP and BIP, 20 pmol of loop primers LF and LB, 1 µl of enzyme mixture containing avian myeloblastosis virus (AMV) reverse transcriptase and Bst DNA polymerase, 12.5 µl of 2X reaction mixture (40 mM Tris-HCl (pH 8.8), 20 mM KCl, 16 mM MgSO4, 20 mM (NH4)2SO4, 0.2% Tween 20, 1.6 M Betaine and 2.8 mM of each dNTPs), 0.12 mM HNB (added before amplification) and 2 µl (~ 100 ng) of target RNA. Reaction was carried out at 61°C for 1 hour and inactivation at 80°C for 5 min in Loopamp real-time turbidimeter (LA-500; Eiken Chemical Co. Ltd., Tokyo, Japan). Positive and negative controls were included in each test and all the precautions were taken to prevent cross-contamination.
Monitoring of Amplification by the RT-LAMP Assay
Real-Time monitoring: Real-time monitoring of rabies viral RNA amplification was done by recording the optical density (OD) at 400 nm at every 6 seconds using Loopamp real-time turbidimeter (Eiken Chemical Co., Ltd; LA-500, Japan). The cutoff value for positive samples was determined by measuring the time of positivity (Tp; in min). The threshold turbidity was fixed at 0.1, which is two times more than the average turbidity value of the negative controls of several replicates.
Agarose gel analysis: Following incubation at 61°C for 60 min, a 5 µl aliquot of the RT-LAMP assay products were electrophoresed on 1.5% agarose gel stained with 0.5 µg/ml ethidium bromide. The amplified products were visualized on a UV transilluminator at 302 nM.
Naked-Eye visualization by HNB dye: Amplification of the RT-LAMP assay was monitored using hydroxynaphthol blue (HNB) dye, through naked-eye inspection. Visual detection of RT-LAMP products was carried out based on colour differentiation between positive and negative reactions using 0.12 mM HNB (Sigma, USA). In case of positive amplification, purple colour of the reaction mixture changed to sky blue, which can be judged under natural light. Original purple colour of the dye indicates absence of amplification. This change of colour is permanent which can be stored for record purposes.
One Step RT-PCR
In order to compare the sensitivity of the RT-LAMP assay, Onestep RT-PCR was performed using a method described earlier by Nagarajan et al. (2009). The amplification was carried out in a 25 µl reaction volume with Onestep RT-PCR kit (Qiagen). The RT-PCR thermal profile was as follows: cDNA synthesis at 50°C for 30 min, initial denaturation at 95°C for 15 min followed by 35 cycles of 95°C for 45 sec, 55°C for 45 sec, 72°C for 1 min, and final extension at 72°C for 15 min.
Analytical Sensitivity of RT-LAMP Assay
Analytical sensitivity of the assay was determined using in-vitro transcribed viral RNA. 605 bp part of N gene of rabies virus was amplified by RT-PCR and the PCR product was cloned into TOPO TA vector (TOPO® TA Cloning®; Thermo Fisher Scientific) downstream T7 promotor. The plasmid clone was confirmed by restriction enzyme analysis and sequencing. N gene RNA was generated from the plasmid clone by in-vitro transcription using TranscriptAid T7 High Yield Transcription kit (Fermentas). The transcribed RNA was purified and stored at -80°C until further use.
Concentration of the RNA was determined by measuring the absorbance at 260 nm in a spectrophotometer (NANODROP 1000; Thermo Fisher Scientific). Copy number of the RNA was calculated using online software (http://scienceprimer.com/copy-number-calculator-for-realtime-pcr). The working standard of positive RNA (in vitro transcribed) was adjusted to 1X1012 copies of N-gene transcripts per 5 µl. A serial ten-fold-dilution was made from the known copy number of in-vitro transcribed RNA (1012 to 100 copy numbers/reaction). RT-LAMP assay and RT-PCR were performed from each dilution of the RNA.
Similarly, ten-fold serial dilutions of cell culture titrated rabies virus was prepared (104 to 10-2 FFU/reaction) and the RNA isolated from each dilution was analysed by RT-LAMP assay. In addition to that, sensitivity of the assay was also evaluated by doing tenfold serial dilutions (100 ng to 10 fg) were prepared using total RNA extracted from representative RABV samples, RV-154 and RV-205.
Analytical Specificity of RT-LAMP Assay
The authenticity of the RT-LAMP assay was ascertained by restriction endonuclease digestion and nucleotide sequencing of amplified products. Restriction digestion of RT-LAMP amplified product was performed using a single restriction enzyme in order to establish the specificity of the amplification product. The suitable unique RE site was determined using an online software (http://creisle.github.io/creisle.lamprflp/). The RE reaction was performed by incubating the RT-LAMP product (2 µl) with 0.5 µl of SphI enzyme (New England Biolabs, MA, USA) at 37°C overnight and the reaction was verified by size comparison on agarose gel electrophoresis. In addition, the amplified products were also confirmed by nucleotide sequencing of both digested and undigested products with two outer and two inner primers. The sequence thus obtained were analyzed and compared with the rabies virus N gene sequence.
Analytical specificity of the rabies RT-LAMP assay was also confirmed by cross-reaction studies using other RNA viruses such as foot-mouth disease virus (FMDV) and bovine viral diarrhea virus (BVDV).
Utility of RT-LAMP Assay
Utility of RT-LAMP assay for the detection of rabies viral RNA from clinical specimens was evaluated by performing the test on extracted RNA from the tissue samples of rabies infected animals. A total of 45 clinical samples were subjected to RT-LAMP assay. The Rabies virus was confirmed from these clinical samples earlier by DFA and RT-PCR. These isolates belonged to two different major genetic lineages of rabies virus viz., Sub-continental and Arctic-like 1 (Nagarajan et al., 2006, 2009; Reddy et al., 2014, 2015). The result of RT-PCR was compared with the RT-LAMP assay which was analyzed by above mentioned three different detection methods.
Results
Successful amplification of the RT-LAMP reaction relies on the specificity of primer sets. The primers were designed from a highly conserved sequence of the rabies viral genome. Following sequence alignment, the mismatches among the strains were taken into consideration while designing the degenerate primer sets.
Detection of RT-LAMP Products
Amplification products of the RT-LAMP assay was monitored by real-time turbidimeter, agarose gel electrophoresis and observing the change in colour due to HNB dye addition.
In the real-time turbidimeter, continuous and gradient amplification of the target sequence was observed. The turbidity remained below 0.1 in no template controls.
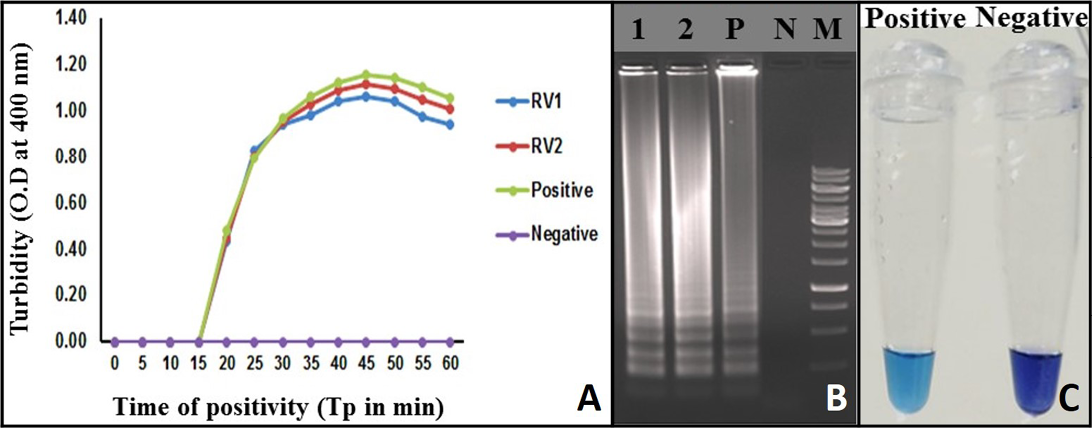
Figure 2: Monitoring of LAMP Amplification: A) Real-time turbidity of RT-LAMP reaction detected by real-time turbidimeter, RV1 and 2: Rabies clinical samples; B) Agarose gel analysis of RT-LAMP amplified products: Lane 1 and 2: Rabies clinical samples; Lane P: Positive; Lane N: Negative control without template RNA; Lane M: 1 Kb DNA ladder; C: Visualization of RT-LAMP products in the presence of HNB dye
Turbidity of the positive samples reached saturation after 58 minutes and showed no further increase. Therefore, a sample with Tp value ≤ 58 and turbidity above the threshold value of ≥ 0.1 was considered positive (Figure 2A).
A typical ladder-like pattern was observed from the positive samples in the agarose gel electrophoresis, indicative of a successful LAMP assay. Such a pattern was not observed, in the negative control (Figure 2B).
The RT-LAMP assay amplification was also monitored by observing a change in the colour of the reaction mixture due to HNB dye addition. The positive RT-LAMP assay was indicated by the sky blue colour of the reaction mix, whereas the negative samples and no RNA template controls retained the original purple colour of the dye (Figure 2C).
Analytical Sensitivity of RT-LAMP Assay
Analytical sensitivity of the RT-LAMP assay was determined using in-vitro transcribed RNA and rabies virus with known titer. Detection limit of the RT-LAMP assay was 103 RNA copies per reaction (Figure 3A). Similarly, the RT-LAMP assay detected up to 10-1 FFU (0.1 Fluorescence focus units) of rabies virus particles per reaction (Figure 3B). The detection limits of RT-LAMP assay and OneStep RT-PCR were similar (Figure 3C). The RT-LAMP assay could able to detect total RNA as low as ~100 fg for both the representative viruses (RV-154 and RV-204), much difference was not observed in Tp values (Tp: Time to positive) (Data not shown).
Analytical Specificity of RT-LAMP Assay
Specificity of the assay was verified by performing digestion with RE enzyme SphI on RT-LAMP product. The SphI has a unique cutting site in the selected target and the resultant digested product was in agreement with the predicted size of 216 bp. Further confirmation of the amplified product was also carried out by sequencing the RT-LAMP product, in which the sequences matched 100% with the reference nucleotide sequences. Analytical specificity of the RT-LAMP was also established by performing the assay using other RNA viruses as template. The genomic RNAs of FMDV and BVDV were used as templates; none of these viruses showed any amplification in the assay. These experiments established the analytical specificity of the assay.
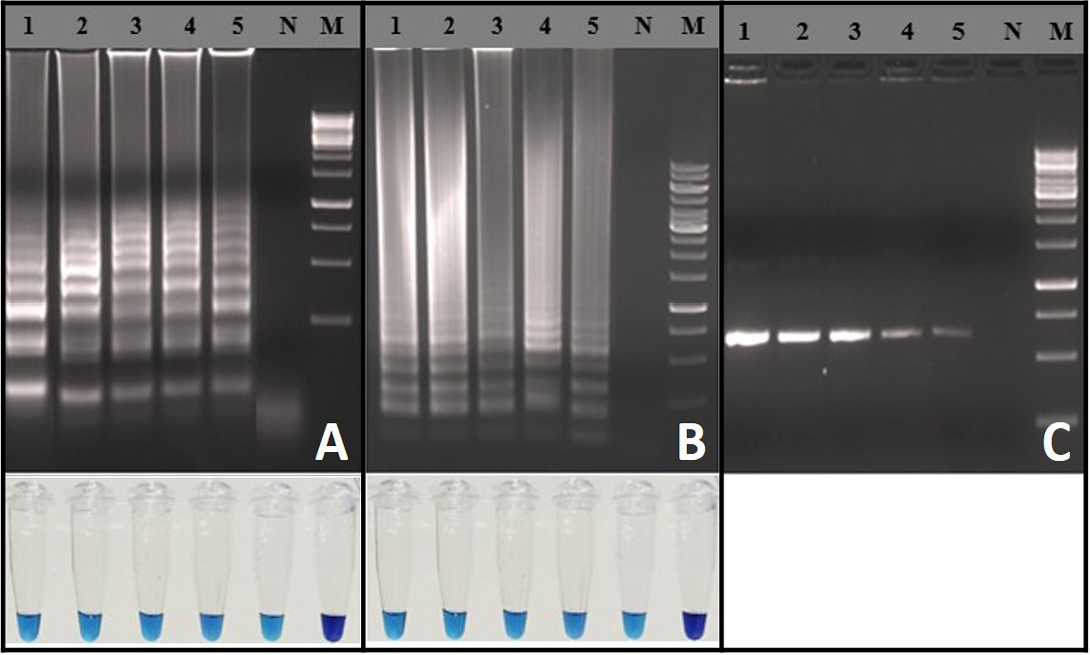
Figure 3: Sensitivity of the RT-LAMP assay: A) Agarose gel electrophoresis analysis with extracted RNA from known FFU of the rabies virus and visual inspection with HNB dye: Lane M: 1 Kb DNA ladder; Lane N: Negative control without template RNA; Lane 1: 104 FFU; Lane 2: 103 FFU; Lane 3: 102 FFU; Lane 4: 101 FFU; Lane 5: 10-1 FFU; B) Agarose gel electrophoresis analysis for copy number determination using in vitro transcribed RNA and visual inspection with HNB dye: Lane M: 100 bp DNA ladder;
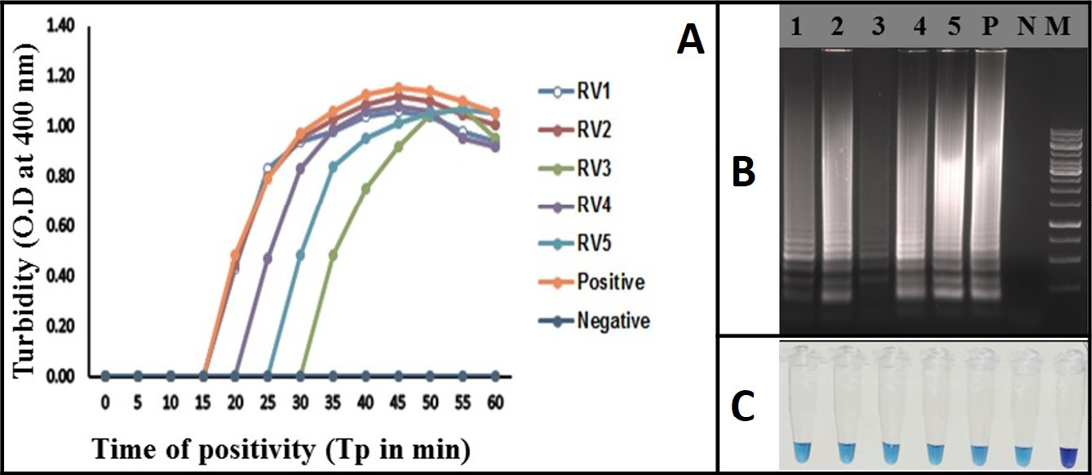
Figure 4: Rabies RT-LAMP evaluation was performed using extracted RNA from infected brain samples. RV1 to RV5 (Lane 1 to 5) are the rabies clinical samples. Negative: Normal-mouse brain tissue; Positive: rabies positive sample; (A) Real-time monitoring of RT-LAMP amplification by turbidimeter; B) Agarose gel analysis revealing the typical electrophoresis pattern of LAMP amplified product; C) Tube containing the amplified RT-LAMP products in the presence of HNB dye
Identification of Rabies Virus Genome from Clinical Samples using the RT-LAMP Assay
The RT-LAMP assay was applied for the detection of rabies virus in clinical samples (brain or/salivary gland tissue). Total of 45 positive samples were tested by the assay. 10 normal mouse brain samples were also used as negative samples. All the RT-PCR positive samples (N=45) were also positive in the RT-LAMP assay and all the 10 normal mouse brain samples turned negative in the assay. All the clinical samples were identified using all the three detection methods of RT-LAMP assay (Figure 4). The diagnostic specificity and sensitivity of the assay was 100% compared to RT-PCR.
Discussion
In the present study, a RT-LAMP assay has been standardized by designing the degenerated primers to detect RABV strains. In particular, while designing the primers N gene sequences of the RABV isolates belonging to major phylogenetic clades defined previously (Nagarajan et al., 2009; Pant et al., 2013; Reddy et al., 2015) were considered. The established RT-LAMP assay showed high degree of specificity as evidenced by detecting all known positive rabies samples of the study belong to three different lineages i.e. Arctic-like 1, sub-continental and cosmopolitan. However, the current assay has not been tested for the other two lineages (Asian and African lineages), due to non-availability of the viral strains. The RT-LAMP assay developed in the current study has facilitated the rapid detection of rabies strains. This test can be conducted in one hour and requires only a heating block or water bath as compared to other conventional test which requires at least a day. This technique is mostly useful for laboratories in developing countries.
During the past decade, various nucleic acid amplification techniques, such as RT-PCR, hemi-nested PCR, TaqMan real-time RT-PCR, and NASBA have been advocated for rabies diagnosis (Heaton et al., 1997; Sugiyama et al., 2003; Nagaraj et al., 2006; Sacramento et al., 1991; Nadin-Davis, 1998; Wacharapluesadee and Hemachudha, 2001; Hughes et al., 2004). These PCR based methods require either high precision instruments for the amplification or elaborate methods for detection of the amplified products. In addition, these methods are often cumbersome to adopt to routine clinical use, especially in peripheral health care settings. In this regard, the RT-LAMP assay of this study is advantageous due to its simple operation, rapid reaction, and easy detection over the other molecular detection methods.
Recently, RT-LAMP assay for the detection of rabies virus have been demonstrated (Boldbaatar et al., 2009; Saitou et al., 2010; Hayman et al., 2011; Muleya et al., 2011). However, in those reported methods designing of RT-LAMP primers was based N gene sequences of only those RABV genetic lineages which were circulating in the particular geographical regions. Boldbaatar et al. (2009) and Hayman et al. (2011) used two different sets of primers to detect the two genetic clusters of rabies virus strains. Saitou et al. (2010) used degenerated bases in loop primers to increase the specificity and sensitivity of the assay. While, the above RT-LAMP methods are more effective for identifying isolates belongs to particular rabies lineages, these methods are probably suitable for the detection of rabies isolates in a particular geographical region. Saitou et al. (2010) demonstrated that the mismatches in the inner primers affect the sensitivity and amplification efficiency of the assay. Thus, in the present study while designing the primers, N gene sequences of different rabies viral lineages and the variations in the sequences were considered and degenerated bases were included.
In the present study, RT-LAMP assay was successfully amplified the rabies viral RNA from all the clinical samples irrespective of the host species and geographical locations with in India. While, the previous RT-LAMP methods for detecting rabies viral RNA extracted from tissue have shown ten folds higher sensitivity compared to the present method (Saitou et al., 2010). This could be due to the inclusion of degenerated bases in the primers. However, the present assay results are in agreement with the previous report from Boldbaatar et al. (2009). Using this assay, rabies virus RNA was detected from the clinical samples and the results were comparable with one step RT-PCR. The assay was performed now using the extracted RNA and further improvement of the assay is needed so that the RNA extraction step can be avoided.
The cost of running the RT-LAMP assay is less expensive than other routine tests such as RT-PCR and FAT. Un-like RT-PCR, RT-LAMP reactions occur at constant temperature. The reaction could be completed in less time and produces large amount of viral nucleic acids that can be visualized with naked eye (Notomi et al., 2000). The incorporation of HNB dye facilitates easy identification by naked-eye, while the earlier methods required separate equipment for visualization (Hayman et al., 2011; Muleya et al., 2011). Therefore, for the field application, HNB dye based detection can be considered as the best method since it is a less expensive and easy to perform point of care detection method.
Conclusion
The present study reports optimization of RT-LAMP-based assay for the detection of rabies viral genome from tissue samples. The sensitivity of the present test was one log lesser compared to the reference rabies RT-LAMP test for the rabies virus detection. The test was performed on field viruses of the Indian rabies isolates and further validation with other field rabies isolates belongs to other lineages may be necessary, this was not attempted as other lineages are not recorded in India. The results are promising for use in peripheral laboratories, where the assay is rapid, easy, simple and sensitive which might be useful for preliminary rabies diagnosis. This will avoid trans-shipment of infective material to well-equipped laboratories. This test does not require sophisticated equipment, which is an essential element in rabies diagnosis in poor resource setting. The method might also facilitate studies of rabies epidemiology where rabies is enzootic, particularly in developing countries.
Acknowledgement
The authors are grateful to the management of the National Diary Development Board (NDDB), Anand for providing the facilities to carry out this work at the Research and Development (R and D) Laboratory, NDDB, Hyderabad. The author RV Chandrasekhar Reddy expresses his gratitude to NDDB and IIL, Hyderabad, for accord the necessary permission for pursuing the doctoral course.
Conflict of interest
There is no conflict of interest.
Authors’ Contribution
Chandrasekhar Reddy R V carried out laboratory work, conducted the experiments, isolated the viruses used in the present study, analysed the data and prepared the manuscript. Krishna Satya A helped in reviewing the data and manuscript. Surendra K S N L helped in designing the laboratory work and analysed the data. RanaS K designed, facilitated the study and finalised the manuscript. Mohana Subramanian Bprovided expert opinions on the design of work and assisted in analysis. SharmaGK helped in facilitating the study, provided the administrative support and facilitated financial support from the NDDB. All the authors are thankful to VA Srinivasan for conceptualisation of the work from time to time, critically reviewed the manuscript and proving a meaningful conclusion from the data generated from the present study.
References