Advances in Animal and Veterinary Sciences
Research Article
Strain Typing of Mycobacterium avium subsp. paratuberculosis from Tamil Nadu, India based on Polymorphisms in MAP 1506 Locus and IS 1311 PCR-REA
Radha Krishna Chaitanya1, Yarabolu Krishna Mohan Reddy2, Arthanari Thangavelu3
1Department of Veterinary Microbiology, College of Veterinary Science, Sri Venkateswara Veterinary University, Tirupati, Andhra Pradesh - 517502, India; 2Vaccine Research Centre-Viral Vaccines, Centre for Animal Health studies, Madhavaram milk colony, Tamil Nadu Veterinary and Animal Sciences University, Tamil Nadu- 600051, India; 3Department of Veterinary Microbiology, Madras Veterinary College, Tamil Nadu Veterinary and Animal Sciences University, Tamil Nadu- 600007, India.
Abstract | Mycobacterium avium subsp. paratuberculosis (MAP) causes Johne’s disease, a progressive chronic granulomatous enteritis affecting the domestic ruminants. Dung samples from cattle, sheep and goat were collected from the suspected clinical cases in livestock farms of different regions of Tamil Nadu state, India. All the samples were subjected to IS 900 PCR for the detection of MAP and 45% were found to be positive. All the positive samples were subjected to IS 1311 PCR-REA that confirmed MAP ‘Bison type’ is most prevalent cause of JD among the domestic ruminants in Tamil Nadu. MAP isolation has been attempted from the tissue samples collected from the animals that were slaughtered being positive for JD. A total of 4 isolates were obtained and were characterized by IS1311 PCR-REA, MAP1506 PCR and sequence analysis which identified the single nucleotide polymorphisms that allow strain typing. One isolate from sheep intestine was identified as “Intermediate type” strain by MAP 1506 PCR-sequence analysis. To the best of our understanding, this is the first report of ‘Intermediate type’ strain of MAP from sheep from India.
Keywords | Mycobacterium avium subsp. paratuberculosis, strain typing, Tamil Nadu, IS 1311 PCR-REA, MAP1506 PCR and sequence analysis.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | December 16, 2014; Revised | January 05, 2015; Accepted | January 06, 2015; Published | April 10, 2015
*Correspondence | Radha Krishna Chaitanya, Sri Venkateswara Veterinary University, Tirupati, Andhra Pradesh, India; Email: chaitanyaerk@gmail.com
Citation | Chaitanya RK, Krishna Mohan Reddy Y, Thangavelu A (2015). Strain typing of Mycobacterium avium subsp. paratuberculosis from Tamil Nadu, India based on polymorphisms in MAP 1506 locus and IS 1311 PCR-REA. Adv. Anim. Vet. Sci. 3(5): 289-294.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.5.289.294
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Chaitanya et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Johne’s disease (JD) caused by Mycobacterium avium subsp. paratuberculosis (MAP) affects wide range of hosts and is responsible for significant economic losses to the livestock industry, due to poor productivity, wasting and early culling of animals. MAP strains have been classified into three groups: Type I (Sheep type), Type II (Cattle type) and Type III (Intermediate type) based on IS 900 PCR- Restriction fragment length polymorphism (RFLP), pulsed field gel electrophoresis (Juan et al., 2005); analysis of sequences of gyr A, gyr B and inh A genes (Castellanos et al., 2007) and based on polymorphisms in MAP 1506 locus (Griffiths et al., 2008). MAP strains isolated from bison in Montana, USA were identified as ‘Bison type” based on polymorphisms in insertion sequence IS 1311 by Whittington et al. (2001) who developed a PCR-Restriction Enzyme Analysis (REA) method that differentiate Cattle, Sheep, and Bison type strains of MAP. MAP strains from India have also been identified as belong to ‘Bison type’ but are different from those of USA and represent a new biotype and have wide host range. Recent reports (Singh et al., 2010; Kaur et al., 2011) have pointed out the existence of ‘cattle type/ Type I’ strains in cattle, buffalo and goats in north India. Also there is evidence of strain sharing and interspecies transmission among different host species (Pavlik et al., 1999; Whittington et al., 2001).
Strain typing can also be achieved by identifying single nucleotide polymorphisms SNPs in MAP 1506 gene locus. MAP 1506 (MACPPE23) is a member of PPE protein family and contains MAP type specific SNPs in its sequence. Proline-Proline-Glutamic acid (PPE) proteins constitute a polymorphic protein family that is restricted to mycobacteria. Previously, these SNPs were targeted to type MAP isolates by Griffiths et al. (2008) and Castellanos et al. (2010). The objective of the present study was to identify the strain types of MAP affecting the domestic ruminant population in Tamil Nadu, India. An IS 1311 PCR- REA, MAP 1506 PCR and sequence analysis were used to differentiate between MAP isolates from cattle, sheep and goats.
Materials and Methods
The study includes a total of 213 dung samples collected from cattle (n=93), sheep (n=68) and goat (n=52) with long term history of emaciation from the organized livestock farms of Tamil Nadu. Intestine samples from cattle (n=4) and a sheep (n=1) were also collected from the animals that were slaughtered after being confirmed as positive for JD. Genomic DNA from the dung and tissue samples was extracted using Quiagen stool DNA mini kit and Quiagen tissue DNA extraction kits, respectively following the manufacturer’s instructions.
Detection of MAP by IS900 PCR
Polymerase chain reaction to amplify a 229 bp product of IS 900 gene was carried out as described by Vary et al. (1990) with few modifications. The reaction mixture (25 µl volume) for IS 900 PCR for each sample include 4µl of DNA template, 12.5 µl of master mix, 1µl each of Forward and Reverse primers (10 pico moles concentration) and 6.5 µl nuclease free water (NFW). The PCR conditions were initial denaturation at 94ºC for 5 min., and 35 cycles of 94ºC for 30 sec., 55ºC for 15 sec., 72ºC for 30 sec., and final extension at 72ºC for 7 min.
IS 1311 PCR – REA for Strain Identification
The samples found positive for MAP by IS 900 PCR were subjected to PCR for amplification of a 608 bp region of IS 1311 gene of MAP and restriction enzyme analysis as described by Marsh et al. (1999).The reaction mixture for IS 1311 PCR includes 3µl of DNA template, 14 µl of master mix, 1µl each of Forward and Reverse primers (10 pico moles concentration) and 6 µl NFW. The PCR conditions were initial denaturation at 94ºC for 5 min and 35 cycles of 94ºC for 30 sec., 62ºC for 15 sec., 72ºC for 60 sec., and final extension at 72ºC for 7 min. The amplicons of IS 1311 gene were subjected to restriction enzyme analysis using Hinf 1 and Mse 1. Reaction mixture for restriction enzyme analysis includes 8 µl of IS 1311 PCR amplicon, 1 µl each of Hinf 1 and Mse 1 , 1.6 µl each of RE Buffer and bovine serum albumin (BSA) which was made up to 16 µl with NFW.
Isolation of MAP and Strain Typing
Isolation was carried out in Middlebrook 7H9 broth and Herrold’s Egg Yolk Medium (HEYM) from all the five tissue samples as per OIE, 2008. The isolates were characterized by IS 900, IS 1311 PCR- REA, MAP1506 PCR and sequence analysis. The sequences of IS 1311 amplicons of all the isolates were analysed for the presence of restriction enzyme sites using NEB cutter.
MAP 1506 PCR
The isolates were characterized by PCR for amplification of 499 bp region of MAP 1506 gene as per the method of Castellanos et al. (2010) and sequence analysis. The sequences were compared with gene sequences of MAP K-10 reference strain and MAA 104 available with the GenBank. Multiple alignment of nucleotide sequence of partially amplified MAP 1506 PCR products was done by CLUSTAL-W method and a phylogenetic tree was drawn by Neighbour-Joining method using MEGA 5 software. The pair wise identity matrix was done in MegAlign software (DNASTAR Inc).
Results
The concentration of DNA extracted from dung samples assessed by Biophotometer Plus (Eppendorf) ranged from 10-25 μg/ml and DNA from tissue samples ranged from 22-35 μg/ml.
Detection of MAP by IS 900 PCR
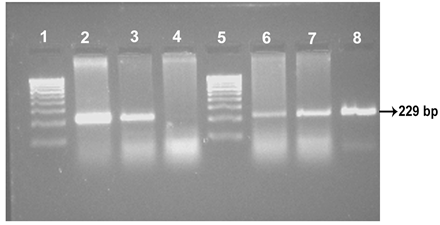
Out of 213 samples of dung subjected to IS 900 PCR, 95 samples (45 from cattle, 32 from sheep and 18 from goat) (45 per cent) revealed specific amplicons of 229 bp size suggestive of MAP infection. All the tissue samples (100 per cent) were found positive for MAP.
IS 1311 PCR-Restriction Enzyme Analysis for Strain Identification
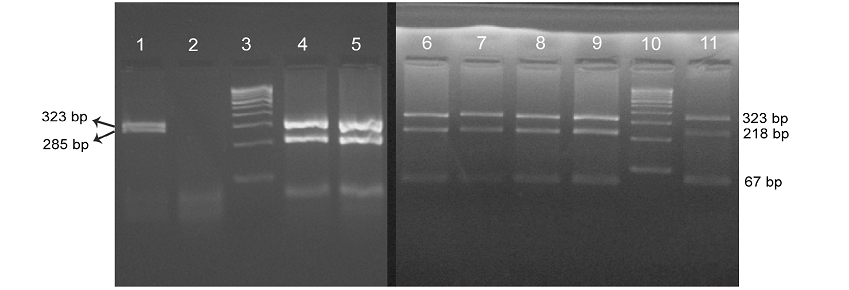
All the samples which were positive by IS 900 PCR were also confirmed for the IS 1311 specific amplicons of 608 bp size. Restriction digestion of amplicons using Hinf 1 and Mse 1 restriction enzymes revealed three bands of sizes 67 bp, 218 bp and 323 bp in all the samples confirming them as ‘bison type’. The intestine sample from Ooty sheep revealed two bands of 323 bp and 285bp.
Isolation of MAP and Strain Typing
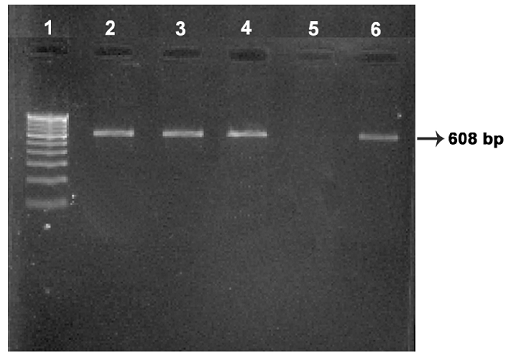
Four isolates viz., Ooty sheep, MVC cattle, MVC goat, URF cattle were obtained in MB 7H9 broth after 12-16 weeks. Growth in culture was confirmed as MAP by Ziehl-Neelsen staining followed by IS 900 PCR (Figure 1). All the four MAP isolates obtained in the present study along with ABT cattle isolate received from the Department of Animal biotechnology, Madras Veterinary College, TANUVAS and BV-I and BV-II sheep isolates received from Bacterial Vaccines division, TANUVAS were characterized. All the seven isolates generated IS 1311 specific amplicons of 608 bp size (Figure 2). Restriction enzyme digestion of IS 1311 amplicons with Hinf 1 and Mse 1 resulted in two fragments of 323 bp and 285 bp for Ooty sheep isolate, a band pattern specific for MAP ‘Sheep type’. All other isolates revealed three fragments of 323 bp, 218 bp and 67 bp, a band pattern specific for ‘Bison type’ strains of MAP upon electrophoresis of RE digested product in 4 per cent agarose gel (Figure 3).
Lane 1 & 5:100 bp DNA marker; Lane 2: ABT Cattle isolate; Lane 3: URF Cattle isolate; Lane 4: Negative control: Lanes 6, 7, 8: MVC Goat, MVC cattle and Ooty sheep isolates, respectively.
Lane 1: 100 bp DNA marker; Lanes 2, 3, 4: Ooty sheep, URF cattle, MVC cattle isolates respectively; Lane 5: Negative control: Lane 6: MVC goat isolate.
Sequence analysis of IS 1311 gene of the isolates in comparison with the MAP K-10 reference strain and MAA 104 complete genome revealed point mutations at base positions 68, 236, 422 and 527 that differentiate these from Mycobacterium avium subsp. avium (MAA) and confirming them as MAP. ABT/MVC/URF cattle, MVC goat, BV-I and II sheep isolates showed an additional point mutation/ nucleotide polymorphism (C/T) at base position 223 that causes a second Hinf 1 restriction site. Whereas the Ooty sheep isolate has ‘C’ at 223 bp position. The sequences were also analysed for the presence of MAP specific restriction enzyme (Hinf 1) sites using NEB cutter (New England Bio Labs online software). Ooty sheep isolate revealed a single Hinf 1 restriction site and ABT isolate had two Hinf 1 restriction sites. None of the isolates shown Mse 1 restriction site which is specific for MAA.
MAP 1506 PCR and Sequence analysis
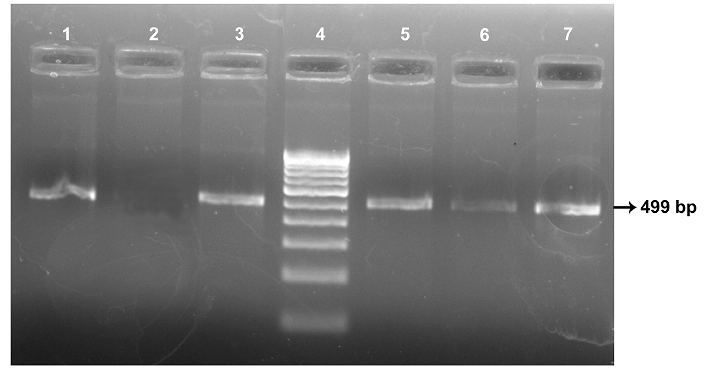
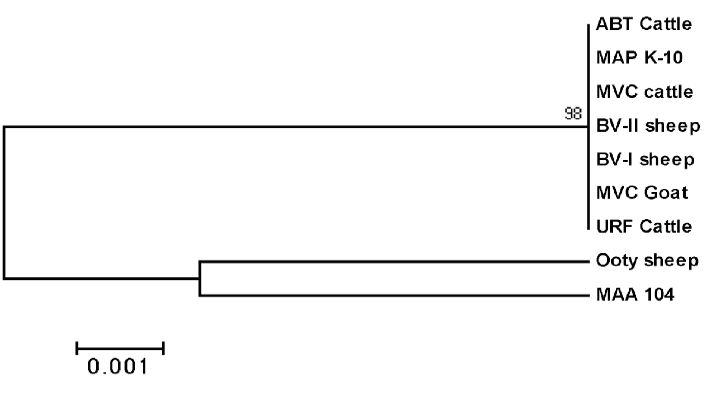
All the seven MAP isolates showed specific bands of 499 bp of MAP 1506 gene (Figure 4). Sequence analysis of MAP 1506 gene loci from all the six isolates (ABT/MVC/URF cattle, MVC goat, BV-I and II sheep isolates that were identified as ‘Bison type’ in IS 1311 PCR-REA and sequence analysis) have nucleotide substitutions (nucleotide 293, 328, 344, 411 and 542) that that categorized them into Cattle/Bison type (Type II) and clearly distinguished them from Type I strains. The Ooty sheep isolate (classified as “Sheep type” by IS 1311 PCR-REA) has SNPs that classified it into ‘Intermediate type’ and these SNPs are also present in MAP 1506 homologues in MAA (Table 1).
Phylogenetic analysis revealed that Ooty sheep isolate was closely related to MAA with percent identity of 99.1%. Remaining isolates are closely related to MAP K10 with the percent identity of 99.8 to 100% (Figure 5). The partial nucleotide sequences of MAP 1506 gene of the isolates were submitted to the GenBank and allotted with the accession number JF 794776 for Ooty sheep isolate and KP 326413 to 326418 for ABT cattle, BV-I, BV-II sheep, MVC cattle, MVC goat and URF cattle isolates respectively.
Discussion
In the present study clinical samples (dung and tissues) were screened by IS 900 PCR as this insertion sequence is found to be specific for this subspecies and being present in 14-18 copies in the MAP genome offers high sensitivity of detection by PCR (Green et al.,1989; Bull et al., 2000; Vansnick et al., 2004).
Lane 3 & 10: 100 bp DNA marker; Lane 1: Ooty sheep showing ‘Sheep type’ IS 1311 band profile with 323 bp and 285 bp band; Lane 4-9: BV-l & ll sheep, ABT/MVC/URF cattle, MVC goat isolates, repectively, showing ‘bison type’ pattern with 323 bp, 218 bp and 67 bp fragments; Lane 11: BV-ll isolate.
Table 1: SNPs identified within the MAP 1506 gene of isolates with different IS 1311 profiles
|
Isolate |
Host animal |
IS1311 PCR-REA Profile |
Position on MAP 1506 gene |
Strain type |
||||
|
bp 293 |
bp 328 |
bp 344 |
bp 411 |
bp 542 |
||||
|
ABT-Cattle |
Cattle |
Bison type |
T |
G |
G |
T |
T |
Type II |
|
MVC-Cattle |
Cattle |
Bison type |
T |
G |
G |
T |
T |
Type II |
|
URF- Cattle |
Cattle |
Bison type |
T |
G |
G |
T |
T |
Type II |
|
MVC-Goat |
Goat |
Bison type |
T |
G |
G |
T |
T |
Type II |
|
BV-I Sheep |
Sheep |
Bison type |
T |
G |
G |
T |
T |
Type II |
|
BV-II Sheep |
Sheep |
Bison type |
T |
G |
G |
T |
T |
Type II |
|
Ooty-Sheep |
Sheep |
Sheep type |
A |
T |
A |
G |
C |
Type III |
|
MAA 104 CP000479.1 |
avium type |
A |
T |
G |
G |
C |
MAA* |
|
|
MAP K-10 AE016958.1 |
Cattle type |
T |
G |
G |
T |
T |
Type II |
|
* Mycobacterium avium subsp. avium
Lane 4: 100 bp DNA marker; Lane 1: ABT cattle isolate; Lane 2: Negative Control; Lane 3: Ooty sheep isolate; Lane 5, 6, 7: MVC goat, MVC cattle and URF cattle isolates, repectively.
Identification of MAP strains prevailing in a geographical region helps in tracking the origin of infection, the risk factors contributing its transmission, pathogenecity and helps in designing the control programmes (Motiwala et al., 2006). Several molecular techniques have been developed by scientists to characterize MAP strains. IS 900-RFLP (Pavlik et al., 1999), Pulsed field gel electrophoresis analysis (Stevenson et al., 2002; Juan et al., 2005) of the genome enable characterization of MAP strains into ‘Sheep/Type I’, ‘Cattle/Type II’ and ‘Intermediate/ Type III’. But these techniques require high quantity of pure genomic DNA and hence difficult to employ for field samples directly and extremely slow growing ‘Sheep type’ strains. Rapid tools like DMC-PCR (Collins et al., 2002) and RDA-PCR (Dohmann et al., 2003) can distinguish MAP Type II (Cattle) strains from Type I (Sheep) strains. However, these techniques cannot further differentiate type III (Intermediate) strains from type I. Moreover, these techniques are unable to detect “Bison Type” strains which have been reported to be the predominant MAP strains causing JD among livestock in North India (Sevilla et al., 2005; Sohal et al., 2009).Keeping in view these facts, in the present study IS 1311 PCR and restriction enzyme analysis using Hinf 1 and Mse 1 which can distinguish MAP strains into Cattle type, Sheep type and Bison type were employed to identify the prevalent MAP strains directly from the clinical samples in Tamil Nadu, India. In the current study, it was found that, in all the cases of JD the strain of MAP involved is of ‘Bison type’ except in one case which was identified as ‘Sheep type’ indicating the heterogeneity of MAP in cattle and sheep.
The results of this study are in accordance with the observations of Sevilla et al. (2005), Singh et al. (2010) and Kaur et al. (2011) who reported that MAP ‘Bison type’ is the predominant strain type affecting livestock population in India. Diversity in MAP strains isolated from cattle and sheep have been reported earlier and were attributed to host-pathogen interactions, specificity or host preference of MAP strains and adaptation to the environmental factors by Whittington et al. (2000) and Bhide et al. (2006).
IS 1311 PCR-REA cannot differentiate type III (intermediate) strains from type I (Sheep type) strains. Hence sequence analysis of MAP 1506 locus that can discriminate between MAP type I and III strains is employed to further characterize the isolates. Griffiths et al. (2008) reported sequence differences in MAP 1506 locus between MAP type I, II and intermediate (type III) strains and developed a PCR- degenerating gradient gel electrophoresis that distinguish these strains. Based on the polymorphisms, Castellanos et al. (2010) developed a real time PCR-High resolution melt analysis targeting 499 bp region of MAP 1506 locus for strain typing.
The present results are in agreement with Castellanos et al. (2010) and Griffiths et al. (2008), and also in concurrence with the results of IS 1311 PCR-REA results. It is found that the isolate from Ooty sheep is of ‘Intermediate type’ and is more closely related to MAA based on SNPs in MAP 1506 locus. This finding is in agreement with the finding of Whittington et al. (1998) who reported that ‘S’ strains are evolutionary intermediates between MAA and MAP cattle strains based on SNPs in IS 1311 gene. The same was proved in this study through the phylogenetic analysis of IS 1311 genes of the isolates. SNPs have been frequently targeted in epidemiological and evolutionary studies to differentiate between closely related species and strains of bacteria. However, strain typing based on SNPs can be more meaningful when they target SNPs in the functional coding regions of the gene, especially those linked with host specificity.
In the present study, when comparing the results of IS 1311 PCR-REA with published data it is evident that “bison type” MAP stains are prevalent and are the major cause of JD among livestock in Tamil Nadu. However, the possibility of existence of other types cannot be overlooked which is evident from the findings of this study. Sheep/ Intermediate strains may be under reported owing to their extremely slow growth rate which precludes their isolation in culture. Further investigations with large number of isolates are required to understand the epidemiological distribution and significance of the MAP types.
Conflict of Interest
The authors declare that they have no conflict of interest related to the work.
Acknowledgements
The authors are thankful to the Vice-Chancellor, Tamil Nadu Veterinary and Animal Sciences University, Chennai for providing the necessary facilities for this work. We are also thankful to the Department of Biotechnology, Madras Veterinary College and Vaccine Research Centre-Bacterial Vaccines, TANUVAS, Chennai Division for providing MAP isolates.
References
249-257. http://dx.doi.org/10.1016/j.vetmic.2004.12.013
J. Clin. Microbiol. 28(5): 933-937.