Advances in Animal and Veterinary Sciences
Short Communication
Isolation and Characterization of Klebsiella pneumoniae from Respiratory Infection of Yak (Bos grunniens), India
Swati Sahay1, Juwar Doley2, Natesan Krithiga1, Gundallahalli Bayyappa Manjunatha Reddy1, Siddanna Sharangouda Patil1, Bibek Ranjan Shome1, Rajeswari Shome1*
1ICAR-National Institute of Veterinary Epidemiology and Disease Informatics (ICAR-NIVEDI), Yelahanka, Bengaluru–560064, India; 2ICAR-National Research Centre on Yak, Dirang-790 101, Arunachal Pradesh, India.
Abstract | Yak (Bos grunniens) is multipurpose long haired ruminant reared by the poor tribal farmers for wool, milk, meat, etc in the high altitudes of Himalayan region of Indian Territory. There is a serious concern about increasing morbidity and mortality of yak due to respiratory diseases. Nasal swab sample was collected from pneumonic yak in Amies charcoal transport media from Arunachal Pradesh, India and processed for bacterial isolation and identification and direct PCR detection in 18h BHI enriched broth sample. Based on biochemical characteristics and multiplex PCR, the culture was identified as K. pneumoniae (isolate No. KP1) and in in-vitro antibiotic sensitivity test, the isolate was resistant to ampicillin. The 16S rRNA sequence analysis and 16S rRNA secondary structure prediction revealed close geographical relatedness to environmental K. pneumoniae PB12 from River Mahananda from East India and K. pneumoniae JPR 9 isolated from soil samples of Assam. This study describes pneumonia in yak due to K. pneumoniae from Arunachal Pradesh, India by direct PCR detection from enriched clinical sample, isolation, PCR and 16S rRNA sequence, phylogenetic and secondary structure relation studies.
Keywords | Isolation, Klebsiella pneumoniae, Pneumonia, 16S sequencing, Yak
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | November 02, 2016; Accepted | November 22, 2016; Published | December 07, 2016
*Correspondence | Rajeswari Shome, ICAR-National Institute of Veterinary Epidemiology and Disease Informatics (NIVEDI), Yelahanka, Bengaluru–560064, India; Email: rajeswarishome@gmail.com
Citation | Sahay S, Doley J, Krithiga N, Reddy GBM, Patil SS, Shome BR, Shome R (2016). Isolation and characterization of Klebsiella pneumoniae from respiratory infection of yak (Bos grunniens), India. Adv. Anim. Vet. Sci. 4(12): 637-641.
DOI | http://dx.doi.org/10.14737/journal.aavs/2016/4.12.637.641
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Sahay et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Yak (Bos grunniens) is domesticated and multipurpose long haired ruminant found throughout the Himalayan region of South Central Asian countries like China, Nepal, India and Tibet and North Asian countries like Russia and Mongolia (Dubal et al., 2013). In India, yak is reared mostly by the poor tribal farmers for wool, milk, leather and meat in the high altitudes of Himalayan region of Indian Territory. Recently there is a growing concern of infectious and non-infectious diseases affecting yaks causing high morbidity and mortality (Bandyopadhyay et al., 2012).
Klebsiella spp. are mostly associated with sepsis, infections of urinary and respiratory tracts, mastitis and meningitis etc. (Brisse and van Duijkeren, 2005). Pulmonary infections due to K. pneumoniae are often characterized by a rapid progressive clinical course complicated by lung abscesses and cause high mortality and morbidity if remain untreated (Cortés et al., 2002). K. pneumoniae has been isolated from bovines, small ruminants (Suguna et al., 2012), camels (Sharma et al., 2015), foals (Boguta et al., 2002) and non-diarrheic yaks (Goswami et al., 2009).
Isolation of bacteria is gold standard diagnostic approach though various serological and molecular diagnostic methods are gaining importance because of high specificity and sensitivity. Among bacterial identifications, 16S rRNA sequence analysis and 16S rRNA secondary structure prediction modeling (Clarridge, 2004) have proven to be a stable and specific molecular marker for the identification of bacteria. This short communication describes pneumonia in yak due to K. pneumoniae from Arunachal Pradesh, India by direct PCR detection from enriched clinical sample, isolation, PCR and sequence phylogeny and secondary structure relation studies.
Female yak of 5 years age suffering from respiratory distress, off feed, pyrexia, severe nasal discharge, depression during peak winter season where temperature ranges from 4°C to 18°C was reported to the institute. The yak belonged to Arunachali breed from a Shyro village of district Tawang, Arunachal Pradesh maintained at altitudes above 9750 feet and not vaccinated against FMD, HS and BQ diseases. Nasal swab sample was collected from pneumonic yak in Amies charcoal transport media (Hi media, India) and airlifted on cold chain to the laboratory within 48 h. After initial enrichment in brain heat infusion (BHI) broth for 18-24 h, sample was streaked onto 7% sheep blood agar (BA) and incubated for 24-36 h at 370C. Pure mucoid colonies observed were purified on Nutrient Agar (NA) later on Mac Conkey’s Lactose agar (MLA) for detection of lactose fermentation.
The culture was examined for gram’s staining, oxidase, catalase, urease, indole, H2S production on triple sugar iron agar (TSI) and sugar fermentation with sucrose, glucose, mannitol, lactose, adonitol, dulcitol, melibiose and esculin (Cruickshank, 1980). DNA was extracted from 18 h BHI enriched broth sample and pure culture by QIAamp DNA mini Kit and quantified by nanodrop. Multiplex genus (Brisse and Verhoef, 2001) and species (Kovtunovych et al., 2003) PCRs were performed for target genes gyrA (441 bp) and ropB (108 bp), respectively. DNA of K. pneumoniae ATCC BAA 1706 and E. coli ATCC 700336 were used as positive and negative control templates, respectively.
In vitro antibiotic sensitivity was carried out using 10 antibiotics (Bauer et al., 1966) [tetracycline (30mcg), cotrimoxazole (25mcg), cefotaxime (30mcg), piperacillin/Tazobactam (100/10mcg), chloramphenicol (30mcg), gentamicin (10mcg), cefepime (30mcg), ampicillin (10mcg), imipenem (10mcg) and meropenem (10mcg)].
The 16S rRNA sequences were aligned with 10 published sequences from India and other neighboring countries (Table 1) to compare phylogenetic relatedness (www.genebee.msu.su.). Similarly, 16S rRNA secondary structure prediction for complimentary region was analysed using Mfold and RNA fold softwares (Zuker, 1989) and the structure with minimum free energy was considered the most stable structure (Hofacker, 2003; Mathews et al., 2004).
Respiratory diseases cause economic losses to the producers in terms of post-infection loss of condition, reduced milk production and inhibited development (Fels-Klerx et al., 2002). The respiratory pathogens/diseases reported in yak are 41% seroprevalence of BHV-1 by virus neutralization test (VNT) and AB-ELISA (Bandyopadhay et al., 2008); bovine viral diarrhoea (BVD) (Mishra et al., 2004) and 36-89% mortality due to Pateurella multocida type I (Pal, 1993). In respiratory infections, it is difficult to arrive at specific diagnosis because of the involvement of large number of pathogens. The pneumonic yak was sampled to rule out the etiology as many of the yaks in the region had similar symptoms. The highly mucoid, non-hemolytic and lactose fermenting pure colonies were observed on NA, BA and MLA media, respectively from the processed sample.
Table 1: List of K. pneumoniae sequences used in the study
|
S. No. |
Strains |
Accession numbers |
Source of strain |
Place |
Minimum Free energy (Kcal/mol) |
|
1 |
K. pneumoniae KP1/yak |
KP866814 |
Yak |
Arunachal Pradesh, India |
-524.4 |
|
2 |
K. pneumoniae QLR-18 |
KM096439 |
Cattle |
China |
-517.7 |
|
3 |
K. pneumoniae AU45 |
EF032681 |
Goat |
Tamil Nadu, India |
-524.9 |
|
4 |
K. pneumoniae -sheep |
AY963633 |
Sheep |
China |
-524.6 |
|
5 |
K. pneumoniae M.D.K.NA1-13 |
JF690874 |
Deer |
China |
-527.7 |
|
6 |
K. pneumoniae LXY |
KM272989 |
Swine |
China |
-517.9 |
|
7 |
K. pneumoniae KP1513 |
KJ746503 |
Mice |
China |
-519.2 |
|
8 |
K. pneumoniae Amm2 |
KJ950284 |
Human |
Cambridge, USA |
-526.8 |
|
9 |
K. pneumoniae PB12 |
KF192506 |
River Mahananda |
West Bengal, India |
-523.9 |
|
10 |
K. pneumoniae JPR9 |
KM083804 |
Soil, East India |
Assam, India |
-516.4 |
|
11 |
K. pneumoniae CF-S9 |
JX680326 |
Waste water |
Odisha, India |
-529.2 |
The culture was Gram negative short rod reacted positive in catalase, urease and sodium citrate tests and negative in indole and H2S production on TSI agar. Based on biochemical tests and multiplex PCR, the culture was identified as K. pneumoniae (isolate no. KP1) (Figure 1). The isolate was resistance to only ampicillin antibiotic.
In the present study, K. pneumoniae was detected in 18 h BHI enriched broth by PCR. The direct detection appears to be of great value in diagnosis and epidemiological mapping of K. pneumoniae as an alternate to time consuming isolation method in difficult geographical terrain where yaks are inhabited. The 16S rRNA (1456 bp) sequences (NCBI accession number KP866814) and RNA secondary structure prediction revealed close geographical relatedness to environmental K. pneumoniae PB12 from River Mahananda from East India and K. pneumoniae JPR 9 isolated from soil samples of Assam. However, sequence divergence was observed with animals isolates from India and other neighboring countries (Figure 2 and 3). There are records of respiratory diseases in yaks in the region but various etiological agents have been implicated other than K. pneumoniae (Mishra et al., 2004).
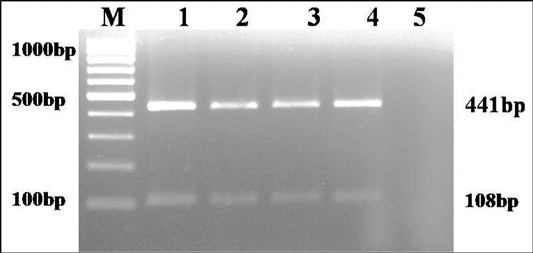
Figure 1: Multiplex PCR amplification of 441bp product of gyrA gene of Klebsiella genus and 108 bp product of ropB gene of K. pneumoniae: Lane M): 100bp molecular marker; Lane1 and 2): 18 hr enriched broth and pure culture DNA of positive control; Lane 3 and 4): PCR amplified products from enriched broth and colony of KP1 cultures; Lane 5): No template control
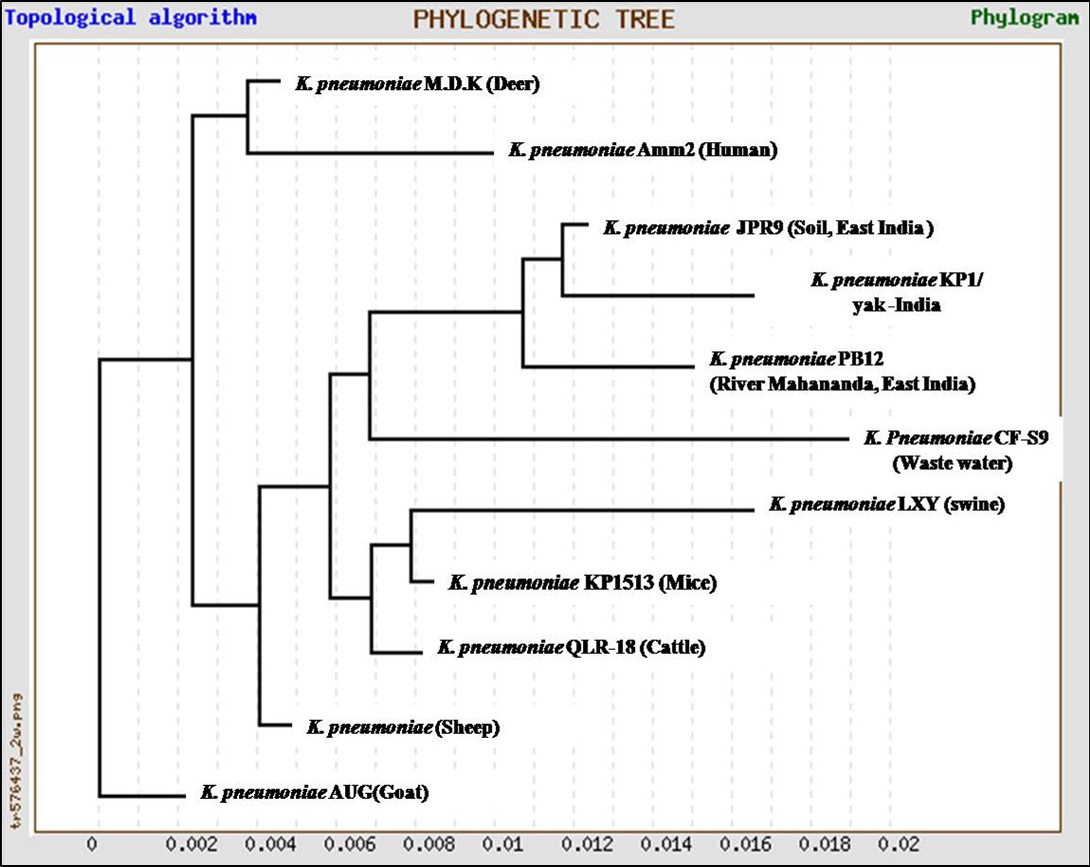
Figure 2: Unrooted phylogenetic tree for 16S rRNA sequences of K.pneumoniae (1372bp) compared with ten environmental and livestock isolates from India and other neighboring countries
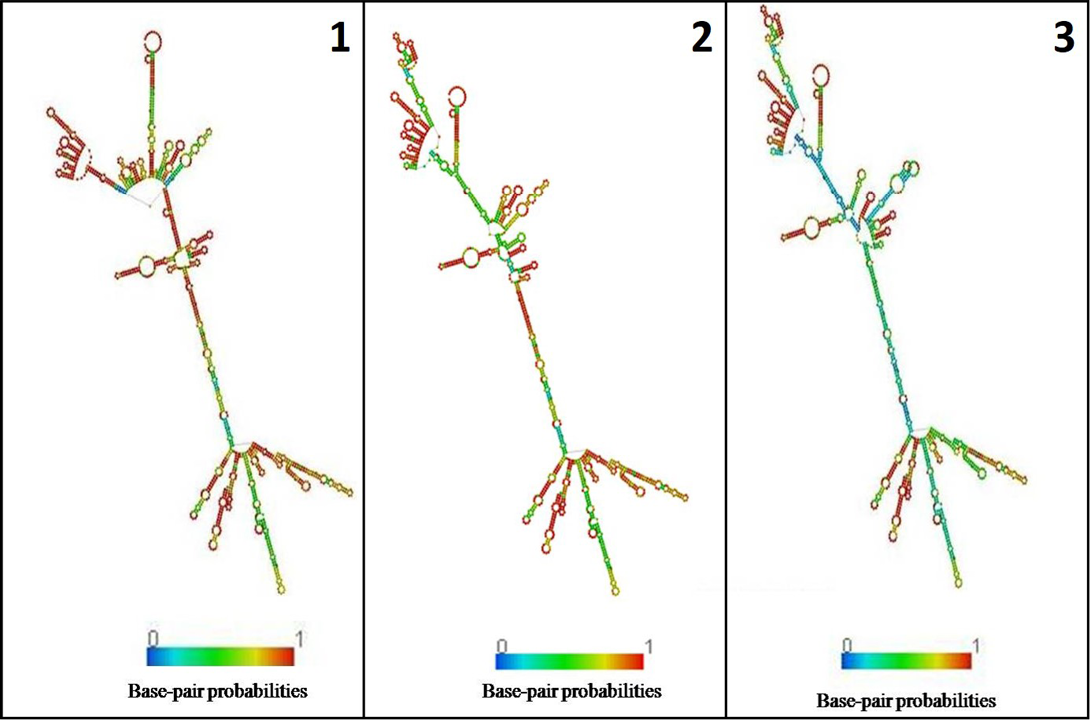
Figure 3: Secondary structure of 16s rRNA: 1) K. pneumoniae yak (524.40 kcal/mol); 2) River Mahananda (523.90kcal/mol); 3) Soil isolates from Assam (516.90kcal/mol)
Identification of K. pneumoniae from respiratory infections reiterates as a respiratory pathogen (Alves et al., 2006; Hansen et al., 2004) and further studies involving large number of isolates and genotyping are essential to understand geographical, pathogencity and physio-chemical factors on the distribution and divergence of K. pneumoniae.
Acknowledgments
We are thankful to the Director and scientists, ICAR-National Research Centre on Yak, Dirang, Arunachal Pradesh for their support & collaboration for sharing samples. This work is funded by Department of Biotechnology Govt. of India through twinning project entitled “Aetio-Pathology and molecular epidemiology of bacterial and viral diseases associated with the respiratory problems of yak in the North Eastern Regions of India (Grant No: BT/391/NE/TBP/2012)”.
Conflict Of Interest
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Authors’ Contribution
RS designed the study and corrected the manuscript. JD provided the sample for study. SS, KN carried out the experimental work, analyzed the data and drafted the manuscript. GBMR,SSP, BRS provided guidance and support. All authors read and approved the final manuscript.
References






