Genetic Characterization of the Endangered Brachymystax lenok tsinlingensis (Salmonidae) Populations from Tsinling Mountains in China using Microsatellite Markers
Genetic Characterization of the Endangered Brachymystax lenok tsinlingensis (Salmonidae) Populations from Tsinling Mountains in China using Microsatellite Markers
Haixia Liu, Yuwei Ye, Yang Li, Xiaolin Liu, Dongmei Xiong* and Lixin Wang
College of Animal Science and Technology, Northwest A&F University, 712100 Yangling, Shaanxi, China
ABSTRACT
Brachymystax lenok tsinlingensis (Salmonidae) is an indigenous and endangered cold-water fish in Tsinling mountain, China, and only is distributed as spot in the Weihe River and Hanjiang River among Tsinling mountain, To access relationship of populations from the two rivers and the genetic diversity of wild population, 11 microsatellite loci were firstly used to evaluate the genetic characterization using 120 samples from 2 tributaries (ZZ and LX populations) of the Weihe river and 2 tributaries (YX and TB population) of Hanjiang river. A total of 59 alleles were found and the number of alleles in each locus ranged from 3 to 8, with an average of 5.4. There were 6 highly polymorphic loci and 4 moderately polymorphic loci. The average expected heterozygosity (He) and observed (Ho) heterozygosity in four populations ranged from 0.6421-0.6980 and from 0.4841-0.6999. Significant deviations from Hardy–Weinberg were found in the four populations at some loci. Analysis of molecular variance (AMOVA) revealed that relatively little (11.02%) genetic diversity came from individual among population. In contrast, the majority of diversity (88.98%) occurred among individual within a population. Results from Nei’s genetic distance indicated that the YX and TB populations were grouped in one cluster, which was clustered with the ZZ population, the LX population was grouped in a separated cluster. Low genetic diversity and strong genetic differentiation have been found in this study. The baseline population genetic information supplied in this study will be vital for the protection and restoration of this endangered species.
Article Information
Received 04 September 2017
Revised 01 November 2017
Accepted 13 November 2017
Available online 26 March 2018
Authors’ Contribution
YY and YL collected the samples and worked on it in lab. XL and LW collected the samples and analyzed the raw experimental data. HL and DX collected the samples and analyzed the experiment results and wrote the paper.
Key words
Brachymystax lenok tsinlingensis, Endangered species, Microsatellite markers, Genetic diversity, Genetic differentiation, Heterozygosity, Polymorphic loci.
DOI: http://dx.doi.org/10.17582/journal.pjz/2018.50.2.743.749
* Corresponding author: liuhaixia209@163.com
0030-9923/2018/0002-0743 $ 9.00/0
Copyright 2018 Zobological Society of Pakistan
Introduction
Brachymystax lenok tsinlingensis (Qinling lenok), as a critically endangered salmonid fish, is only found in Weihe and Hanjing Rivers tributaries(cold freshwater) in Tsinling Mountains, China (Sze-Chung, 1966). In the past few decades, the populations of B. lenok tsinlingensis have been declining dramatically because of illegal overfishing and environment changes (Ren and Lang, 2004). In 1988, it has been list as a second class state protected wild animal in China Red Data Book of Endangered Animal (Yang et al., 1999). It is very urgent for researchers to protect this rare cold water salmonid fish resource with effective methods. Microsatellite is one of the molecular markers that are very useful tools to estimate genetic diversity, evolutionary potential, population structure of the endangered species population (Shao et al., 2017; Was et al., 2002; Liu et al., 2014).
The Tsinling Mountain is an east-west trending south vergent mountain, which has been recognized as the geo-ecological boundary between subtropical and warm-temperate zones. B. lenok tsilinlingensis is one of two Salmonidae located in the southernmost part of China (the other is hucho taimen) (Xia et al., 2016; Wang et al., 2016). There is no information about the distribution characteristics and genetic diversity of B. lenok tsilinlingensis at the northern (Weihe River) and southern (Hanjiang River) of Tsinling Mountain. So, this research aimed at attracting much attention to its genetic diversity, population genetic and the genetic variation assessment of B. lenok tsilingensis in the two river systems.
Material and methods
Sample collection and DNA extraction
One hundred and twenty samples of fin tissues were collected from 4 different tributaries in the northern and southern of Tsinling Mountain between May and September 2015 (Fig. 1, Table I). The fin clips were gathered and stored in 95% ethanol until extraction, then these fish were immediately set back to the rivers.
Genomic DNA was extracted from samples of fish fin tissue using a standard traditional phenol-chloroform procedure (Sambrook and Fritsch, 1989) and standardized to a concentration of 50ng/μl and stored in ddH2O.
Methods
Twenty microsatellite loci were used for the design of the multiplex PCR. The 8 of 18 microsatellite markers were developed in our laboratory using the method of fast isolation by AFLP of sequences containing repeats (FISCO) and primers were designed using Primer 5.0. The other 10 polymorphic primers pairs came from known microsatellite makers in Brachymytax lenok and Hucho bleekeri (Jia et al., 2008). All the primers were synthesized by Nanjing GenScript Biological Engineering & Technology and Service Co. Ltd., Nanjing, China.
For initial testing, the PCR amplifications were carried out in a 10 μl reaction volumes containing the following components: 20 ng of genomic DNA, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.25 μM each primer, 1×Taq buffer and 0.5 U Taq polymerase (Takara). PCR conditions were as follows: initial denaturation at 94°C for 5 min followed by 35 cycles of denaturation 30 s at 94°C, 30 s at a locus-specific annealing temperature (Table II), and extension 30 s at 72°C, with a final extension at 72°C for 8 min. The PCR amplification produces were separated using 8% non-denaturating polypropylene gel electrophoresis (120V, 4°C for 3-5 h), and then the gel was silver stained and scanned. Only 11 primer pairs that could amplify polymorphism were selected for the further study. Amplification conditions were further optimized by adjusting the relative concentration of ingredient and annealing temperatures.
Table I.- Sampling site and date of B. lenok tsinlingensis population.
| Population | Location |
n |
Date |
| Yangxian (YX) |
Xushui river in Yangxian town at the southern foot of Qinling Mountain E 107°17'-107°55'/N 33°19'-33°44'; Elevation 800-3071 m |
40 |
Mar, 2015 |
| LongXian (LX) |
Qianhe river in Longxian town at the northern foot of Qinling Mountain E106°26'-107°06'/N34°35'-35°08'; Elevation 1100-2466 m |
30 |
Sep, 2015 |
|
Taibai (TB) |
Taibai river at huangpoyuan of Taibai town in Baoji Provinces E107°16'-107°42'/N33°38'-33°54'; Elevation 902-1510 m |
30 |
Apr, 2015 |
| Zhouzhi (ZZ) |
Heihe river in Zhouzhi town at the northern foot of Qinling Mountain E107°39'-108°19'/N33°41'-33°57'; Elevation 2996 m |
20 |
Apr, 2015 |
Data analysis
POPGENE32 software was used to estimate the expected heterozygosity:
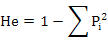
Where, Pi is the frequency of the ith allele (Nei, 1973) and observed heterozygosity (Ho=Na/n; Where Na is the number of heterozygous genotypes, n is the total number of genotypes), fixation index (Fis = 1 - Ho/He (Wright, 1965)), genetic distance (D) among population, genetic differentiation index (Fst), and gene flow (Nm). We tested the linkage disequibrium (LD) using GENEPOP program (online version through http://genepop.curtin.edu.au/) and default parameters. The Markov chain method (Guo and Thompson, 1992) was used to estimate the probability of significant deviation from Hardy-Weinberg equilibrium (HWE).AMOVA was used to analyze the genetic differentiation among the populations. The micro-check software (Van Oosterhout et al., 2004) was used to infer the most probable technical cause of HWE departures, including null alleles, mis-scoring and allelic dropout. Polymorphism information content (PIC) (Bostein D et al., 1980) of each locus was analyzed using the following formula:
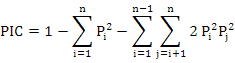
Where, Pi and Pj are allele′s frequencies and n are the locus number.
Table II.- The sequences of 11 pairs of microsatellite primers.
| Locus |
GenBank Accession No. |
Annealing primer (5′-3′) |
Product size (bp) |
Anne aling temp (°C) |
| GTL05 |
JN835587 |
TACATTTCAGTTTGAATCACGCAAC CGTGATGCAGCCCTGCAT |
114-150 |
54.8 |
| GTL08 |
JN835588 |
AGGAAATGTTAGGATGATAGAGGCA TTGGTTATTTAATGCGGTTGGG |
100-140 |
50.0 |
| GTL10 |
JN835590 |
AGCCTACCTCTTCTGTCTAGTGAGG TGTGCAAATAGTTCAAGAACAAAAG |
120-150 |
63.0 |
| GTL12 |
JN835591 |
CTGCAGACTGGATCTTATCAGGAGC GCATACAAGTACGCACGCCGA |
170-200 |
63.0 |
| GTL14 |
JN835592 |
AGGCAGACTAACTGTGTGTATC AGTTAGTGTGAACTCTGTTGTGT |
160-190 |
50.0 |
| GTL15 |
JN835586 |
AGCGACAGTGTGTGTGAGTAAGTGG CCTCAAACCTGATGACCTCACACA |
120-183 |
55.0 |
| GTL21 |
JN835594 |
GAGATTAGATTGGGAGACTATGGTG ATGGTTGTGTTCTCACTCTCTCTTT |
120-140 |
63.0 |
| GTL23 |
JN835595 |
TCCAGTGAGACCTGCCCAGTAACAT ATTTGTGCATGAGTGCAGCATGTG |
128-210 |
53.0 |
| HLJZ069* |
DQ120978 |
ACCGACACAACACAAACA ATGAATGCGGGATAAAGT |
148–165 |
45.0 |
| LETET2* |
AY84452 |
TGTCAGAGGCCTTGACTGCGT GCTAGGCTGTTTACTCTAGGT |
150-169 |
60.0 |
| BLETET9** |
AY486103 |
ACTGGATAGAAAGACCTGTGG AGATTCTTGGTAAAAGTGAAG |
170-230 |
55.0 |
*, microsatellite marker of Brachymystax lenok; **, microsatellite marker of Hucho bleekeri.
Table III.- The genetic diversity indices of eleven microsatellite loci for four Brachymystax lenok tsinlingensis populations.
|
Locus |
ZZ |
TB |
YX |
LX |
||||||||||||
|
Ne |
Ho |
He |
PIC |
Ne |
Ho |
He |
PIC |
Ne |
Ho |
He |
PIC |
Ne |
Ho |
He |
PIC |
|
| GTL05 |
3.88 |
0.70 |
0.74 |
0.65 |
3.32 |
0.41 |
0.67 |
0.59 |
4.19 |
0.91 |
0.76 |
0.68 |
3.74 |
0.55 |
0.73 |
0.64 |
| GTL08 |
4.48 |
1.00 |
0.74 |
0.65 |
3.81 |
0.91 |
0.72 |
0.64 |
4.50 |
1.00 |
0.78 |
0.70 |
3.44 |
0.44 |
0.69 |
0.61 |
| GTL10 |
3.16 |
0.60 |
0.65 |
0.57 |
4.34 |
0.83 |
0.77 |
0.69 |
4.61 |
0.83 |
0.78 |
0.72 |
4.87 |
0.66 |
0.81 |
0.73 |
| GTL12 |
3.03 |
0.80 |
0.63 |
0.52 |
4.19 |
0.91 |
0.76 |
0.68 |
3.16 |
0.83 |
0.65 |
0.54 |
4.02 |
0.66 |
0.75 |
0.67 |
| GTL14 |
3.62 |
0.60 |
0.71 |
0.64 |
3.46 |
0.66 |
0.69 |
0.58 |
3.35 |
0.83 |
0.67 |
0.59 |
4.10 |
0.66 |
0.76 |
0.67 |
| GTL15 |
2.76 |
0.50 |
0.58 |
0.49 |
4.34 |
0.58 |
0.77 |
0.69 |
3.14 |
0.53 |
0.64 |
0.56 |
2.81 |
0.51 |
0.60 |
0.49 |
| GTL21 |
3.44 |
0.40 |
0.69 |
0.61 |
3.43 |
0.66 |
0.68 |
0.63 |
3.46 |
0.75 |
0.69 |
0.61 |
3.15 |
0.33 |
0.66 |
0.55 |
| GTL23 |
2.88 |
0.67 |
0.61 |
0.49 |
2.50 |
0.55 |
0.52 |
0.37 |
2.66 |
0.83 |
0.56 |
0.43 |
2.50 |
0.65 |
0.52 |
0.36 |
| HLJZ069 |
4.27 |
0,70 |
0.77 |
0.68 |
4.01 |
0.83 |
0.74 |
0.66 |
3.40 |
0.58 |
0.68 |
0.58 |
3.50 |
0.33 |
0.70 |
0.60 |
| LETET2 |
2.56 |
0.10 |
0.54 |
0.42 |
2.84 |
0.25 |
0.59 |
0.51 |
3.46 |
0.50 |
0.69 |
0.58 |
2.02 |
0.00 |
0.36 |
0.29 |
| BLETET9 |
2.01 |
0.01 |
0.35 |
0.31 |
3.77 |
0.83 |
0.72 |
0.63 |
3.04 |
0.08 |
0.64 |
0.54 |
3.15 |
0,11 |
0.66 |
0.58 |
|
Mean |
3.23 |
0.53 |
0.64 |
0.55 |
3.64 |
0.67 |
0.69 |
0.61 |
3.54 |
0.69 |
0.68 |
0.59 |
3.39 |
0.48 |
0.66 |
0.56 |
Ne, expected alleles; Ho, observed hererozygosity; He, expected heterozygosity; PIC, polymorphism information contents.
Results
Genetic diversity of analysis
Eleven pairs of polymorphic microsatellite primers were used to analyze genetic diversity of these populations from 4 tributaries. All parameters for genetic variation of each population are summarized in Table III. We amplified a total of 59 polymorphisms loci, the number of alleles per locus ranged from 3-8, with an average of 5.4. The expected (He) and observed (Ho) hererozygosities in the four populations ranged from 0.6421 to 0.6980 and from 0.4814 to 0.6999, respectively. All 11 microsatellite loci were found to be highly polymorphic in four populations, the average of PIC were 0.5527~0.6103.
Hardy–Weinberg equilibrium
The Hardy-Weinberg test on multi-locus based on the Markov chain method showed that there were separately 3, 2, 3, 4 loci deviated from HWE(p<0.01)in each of the four populations, the LX population had the highest level of deviation (four loci). Conversely, the TB population had the least deviation (two loci) The MICRO-CHECKER analysis showed no evidence for scoring error or technical or statistical artifacts, the results of the HWE tests for the individual populations were shown in Table IV.
| Locus |
ZZ |
TB |
YX |
LX |
| GTL05 |
0.00007 |
0.15613 |
0.00102 |
0.04752 |
| GTL08 |
0.57261 |
0.00005 |
0.03502 |
0.09367 |
| GTL10 |
0.03363 |
0.02978 |
0.11302 |
0.00265 |
| GTL12 |
0.01353 |
0.27674 |
0.00001 |
0.03678 |
| GTL14 |
0.00181 |
0.29593 |
0.42283 |
0.00575 |
| GTL15 |
0.06548 |
0.09526 |
0.05202 |
0.05437 |
| GTL21 |
0.07053 |
0.05277 |
0.06116 |
0.07627 |
| GTL23 |
0.02929 |
0.02691 |
0.18728 |
0.01468 |
| HLJZ069 |
0.05442 |
0.41198 |
0.19910 |
0.09557 |
| LETET2 |
0.01059 |
0.00410 |
0.01330 |
0.00071 |
| BLETET9 |
0.00000 |
0.04234 |
0.00000 |
0.00003 |
Note: P < 0.01 denotes Hardy-Weinberg disequilibrium.
Genetic structure of four B. lenok tsinlingensis populations
Gametal correlation coefficient (FST) and Gene Flow (Nm) were computed to estimate the differences between populations. The average of Fst was 0.1109, and the average of Nm among populations was 2.4308 at locus (Table V). It indicated genetic variations within the populations were higher than those among the populations, thus genetic flow might be the significant factor between populations (Table VI, Fig. 2). The AMOVA method was used to analysis the genetic structure of the populations, the result showed that 11.02% of the genetic diversity was distributed among the populations and 88.98% occurred among individuals within the populations.
Table V.- The results of F-statistics for each of 11 loci across four B. lenok tsinlingensis populations.
| Locus |
Fis |
Fit |
Fst |
Nm* |
| GTL05 |
0.0658 |
0.1424 |
0.0819 |
2.8024 |
| GTL08 |
-0.1947 |
-0.0808 |
0.0953 |
2.3728 |
| GTL10 |
-0.0139 |
0.0532 |
0.0661 |
3.5306 |
| GTL12 |
-0.2024 |
-0.0464 |
0.1297 |
1.6775 |
| GTL14 |
-0.0191 |
0.0406 |
0.0587 |
4.0119 |
| GTL15 |
0.2130 |
0.2324 |
0.1193 |
1.5150 |
| GTL21 |
0.1751 |
0.2709 |
0.1161 |
1.9031 |
| GTL23 |
-0.2097 |
-0.2383 |
0.0063 |
5.1083 |
| HLJZ069 |
0.1165 |
0.2122 |
0.1082 |
2.0597 |
| LETET2 |
0.5946 |
0.6435 |
0.2141 |
0.6179 |
| BLETET9 |
0.5463 |
0.6435 |
0.2141 |
0.6179 |
| Mean |
0.0974 |
0.1702 |
0.1109 |
2.0042 |
Table VI.- Analysis of molecular variance (AMOVA) within and among the four B. lenok tsinlingensis populations.
| Source of variation |
df |
Variance component |
Percentage of variation |
FSt |
| Among populations |
4 |
0.8166 |
11.02 |
0.1102 |
| Within individuals |
120 |
6.4322 |
88.98 |
Clustering analysis
Based on Nei’ genetic distance and phylogenetic tree, the highest divergence was between the TB and LX populations (D=0.2640), while the lowest divergence was between TB and YX populations (D=0.0014) which all located at the foot of the south of Tsinling mountains (Table VII).
Table VII.- Nei’s genetic distance of the 4 B. lenok tsinlingensis populations.
|
ZZ |
TB |
YX |
LX |
|
| ZZ |
**** |
|
|
|
| TB |
0.0806 |
**** |
|
|
| YX |
0.1232 |
0.0014 |
**** |
|
| LX |
0.1760 |
0.2640 |
0.1522 |
**** |
Discussion
Results of primer amplification
B. lenok tsinlingensis is endemic to cold-water of Tsinling Mountains and mainly distributed Weihe River and Hanjing River drainage, China. Hanjiang River drainage is southernmost point of B. lenok tsinlingensis distribution (the other Salmonidae that distributed southernmost is hucho bleekeri). However, the adaptive characters and genetic diversity of these populations have been little studied. We analyzed the genetic diversity using 18 microsatellite primers (Liu et al., 2013; Lu et al., 2012), but only 3 of 10 microsatellite markers from Hucho bleekeri and Brachymystax lenok could amplify polymorphic loci in B. lenok tsinlingensis. The results showed that the markers obtained from other salmonids are suitable for study of Qinling lenok (Kuang et al., 2009), but have relatively low availability.
Hardy-Weinberg and linkage disequilibrium
The global Hardy–Weinberg equilibrium (HWE) of all populations and loci were evaluated, and significant departures from Hardy–Weinberg equilibrium were found for several loci in some locations. The four populations had 3, 2 3 and 4 locus deviation after Bonferroni correction (P<0.01), respectively. Processes causing this non-equilibrium were specific to those populations; those loci did not show consistent deviation cross all locations. Main reasons of deviated HEW were attributed to geography and shrinking of population size. The result also was proved by the other landlocked freshwater Salmondae fish such as Hucho taimen (Kuang et al., 2009) and Oncorhynchus keta walbaum (Jinping et al., 2004) in China.
B. lenok tsinlingensis wild population only distributed the tributaries of Weihe and Hanjing River, which separately located the northern and southern of Tsinling Mountain. The distribution areas of the fish have suffered severe effected even vanish due to local water temperature increasing and excessive fishing in the past forty years, Ren and Lang (2004) once reported that B. lenok tsinlingensis population in Qianhe River was not only sharp decrease in number, but also change geographical conditions from altitude 1000 to 1200 m. Thus, the fluctuation of the genotype and gene frequencies is likely due to human influence.
Genetic diversity
The four B. lenok tisinlingensis populations exhibited high genetic diversity (He>0.5and PIC>0.5). Dewoody and Avise (2000) computed nearly 40,000 individuals of 78 species freshwater, marine fish and other animal with 524 microsatellite markers, and suggested genetic diversity of freshwater fish (He=0.58 and Ae=7.1). In the study, the average He here was 0.67 and Ae was 5.4. The He was larger, but Ae was smaller. The reason for accounting for the low Ae is that habitat of these population is in deep water and not be disturb for a long time. Bostein et al. (1980) indicated that the locus was high polymorphic when PIC > 0.50, mediate polymorphic when the PIC was between 0.25 and 0.50. The average PIC in the four population here was 0.5642 - 0.6103, which was higher than the polymorphic level, so the four populations were high PIC population. All this indices were higher than study of Yuan and Wang (2009) that suggested lower genetic diversity in the Heihe and Xushui Rivers populations using PAPD methods, the variance maybe was connected with the population size of different tributaries. Xushui River is the first location where B. lenok tsilinlingensis were migrated and bred, but the number of the parental fish in Xushui River is relatively few and the germplasm resources were deteriorated owing to no scientific breeding.
Relation and genetic differentiations between populations
There was no significant difference (p>0.01) in the four populations. The average of Fst of all loci was 0.1109, which was larger than the standard of no differentiation between populations suggested by Wright (1978). According to the expressions of Nm = (1−Fst) / 4 FST, the average of Nm between populations was 2.0042, the differentiations that were caused by genetic drift were stopped among populations. During the Quaternary glaciations (the period from 2.58 million years ago to present), the B. lenok originated from Siberia has been flowed to the Yellow sea and the Bohai sea with the role of glacial advance, they ran upstream of the Yellow River to Wei River to spawn as they need breed (Xing et al., 2015). Most of the populations backed to primary source as the deglaciation, whereas part of them were stagnated in the high mountain and ravine stream of Wei River basin, and they were sealed off in those regions. After that, the rivers lost the connectivity is aroused by the expansion of human activities and the construction of many engineering structures, and the blocking of the migration route of B. lenok leaded to the point distribution along Weihe River basin, Heredity and aberrance are necessary to adapt to the changing environment, after long time selection, B. lenok tisilingensis, as a subspecies of B. lenok, survived in the Tsinling Mountains (Yu and Kwak, 2015).
The high genetic identities and low genetic differential of the four populations suggested that B. lenok tisilingensis of Weihe River and Hanjiang River derived from the same ancestor. It is said that there was no B. lenok tsinlingensis in the southern of Tsinling Mountain before long years ago, and in the 1980s, the fish were migrated to the tributaries of Yangtze River by the local peasant in Zhouzhi town, these isolation habitats induced changes in population genetic structure. The date (Nm>1) suggested that there was gene flow within four populations.
Population genetic research is important to understand population genetic distance and potential species differentiation (Matocq et al., 2001). YX and TB populations share the highest genetic identity, indicating the nearest relationship. Populations LX and TB had the lowest genetic identity and the relationship was the farthest. Geographic isolation for a long time and genetic variation help them to adapt to their habitats in the course of living and evolution, thus, these population that live close to each other have higher genetic identities and geographic proximity by the effect of gene flow. The results agreed with Brunner et al. (2001) suggestion that Salvelinus alpinus among different geographic population showed the significant genetic variation because of its breeding migration and low distribution.
Despite the decline in the fish population size, the genetic diversity of this species is relatively higher than B. lenok in other basin in China (Xia et al., 2006). B. lenok tsinglingensis only distributed in cold-water mountain streams and rivers with elevations ranging from 900-2,300 m above sea level. Though environmental change by dam construction and mining led to the shrinking geographical distribution, the spawning and water source remain healthy due to high altitude, the low density and the discontinuous distribution. In recent year, the nature protected areas of B. lenok tsinglingensis have been established by local government to protect the germplasm resources.
In summary, though shrinking geographical distribution of B. lenok tsinlingensis, there is high level of genetic diversity in four locations. However, limited number of alleles and deteriotate of habitat may lead to deviate from HWE, and then genetic diversity will sharply decline. Thus, we should be protected the fish from further declines in genetic diversity.
Acknowledgments
This research was financially supported by Fundamental Research Funds for the Central Universities (Z109021512) and National Natural Science Foundation of China (31702344 and 31302189).
Statement of conflict of interest
The author(s) declare(s) that there is no conflict of interests regarding the publication of this article.
References
Brunner, P.C., Douglas, M.R., Osinov, A., Wilson, C.C. and Bernatchez, L., 2001. Holarctic phylogeography of Arctic charr (Salvelinus alpinus L.) inferred from mitochondrial DNA sequences. Evolution, 55: 573-586. https://doi.org/10.1554/0014-3820(2001)055[0573:HPOACS]2.0.CO;2
DeWoody, J.A. and Avise, J.C., 2000. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol., 56: 461-473. https://doi.org/10.1111/j.1095-8649.2000.tb00748.x
Guo, S.W. and Thompson, E., 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics, 2: 361-372. https://doi.org/10.2307/2532296
Jia, Z.Y., Zhang, Y.Y. and Shi, L.Y., 2008. Amplification of rainbow trout microsatellites in Brachymystax lenok. Mol. Ecol. Resour., 8: 1520-1521. https://doi.org/10.1111/j.1755-0998.2008.02310.x
Kuang, Y.Y., Tong, G.X., Xu, W., Yin, J.S. and Sun, X.W., 2009. Analysis of genetic diversity in the endangered Hucho taimen from China. Acta Ecol. Sin., 29: 92-97. https://doi.org/10.1016/j.chnaes.2009.05.002
Liu, H.X., Li, Y., Liu, X.L., Zou, G.W. and Wei, Q.W., 2013. Isolation and characterization of eleven novel microsatellite loci of Brachymystax lenok tsinlingensis, a threatened fish endemic to Shaanxi, China. Conserv. Genet. Resour., 5: 389-391. https://doi.org/10.1007/s12686-012-9810-7
Liu, D.Q., Wu, J. Y., Deng, L.J., 2014. Development of microsatellite markers for Leptobotia elongata (Cypriniformes: Cobitidae) using 454 sequencing and cross-species amplification. Pakistan J. Zool., 46:1147-1151.
Lu, X., Wang, H., Liu, B. and Lin, Z., 2012. Microsatellite-based genetic and growth analysis for a diallel mating design of two stocks of the clam, Meretrix meretrix. Aquacul. Res., 43: 260-270. https://doi.org/10.1111/j.1365-2109.2011.02823.x
Matocq, M.D. and Villablanca, F.X., 2001. Low genetic diversity in an endangered species: Recent or historic pattern? Biol. Conserv., 98: 61-68. https://doi.org/10.1016/S0006-3207(00)00142-7
Ren, J. and Lang, G., 2004. Resource survey report of Brachumystax lenok tsilingensis in Qinhe river valleys of Tsinling Mountains. J. Shaanxi Norm. Univ. (Nat. Sci. Ed.), 32: 165-168.
Sambrook, J. and Fritsch, E., 1989. Molecular cloning. Cold Spring Harbor Laboratory Press, New York.
Shao, J., Luo, W., Wei, Q.W., Wang, F., Guo, W., Ye, H., Chu, Z.P. and Wu, J.M., Zhang, SH., 2017. Assignment of parentage by microsatellite analysis in the endangered Brachymystax lenok tsinlingensis (Salmonidae). Aquat. Biol., 26: 69-73. https://doi.org/10.3354/ab00675
Van Oosterhout, C., Hutchinson, W.F. and Wills, D.P.M., 2004. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Resour., 4: 535-538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Wang, K., Zhang, S.H., Wang, D.Q., Wu, J.M., Wang, C.Y. and Wei, Q.W., 2016. Conservation genetics assessment and phylogenetic relationships of critically endangered Hucho bleekeri in China. J. appl. Ichthyol., 32: 343-349. https://doi.org/10.1111/jai.13018
Was, A. and Wenne, R., 2002. Genetic differentiation in hatchery and wild sea trout (Salmo trutta) in the Southern Baltic at microsatellite loci. Aquaculture, 204: 493-506. https://doi.org/10.1016/S0044-8486(01)00835-3
Wright, S., 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution, 19: 395-420.
Xia, J.G., Ma, Y.J., Fu, C., Fu, S.J. and Cooke, S.J., 2016. Effects of temperature acclimation on the critical thermal limits and swimming performance of Brachymystax lenok tsinlingensis: A threatened fish in Tsinling Mountain region of China. Ecol. Res., 32: 61-70. https://doi.org/10.1007/s11284-016-1418-z
Xia, Y., Yan, S. and Yiyu, C., 2006. DNA sequence variation in the mitochondrial control region of lenok (Brachymystax lenok) populations in China. Biodiv. Sci., 14: 48-54. https://doi.org/10.1360/biodiv.050189
Xing, Y.C., Lv, B.B., Ye, E.Q., Fan, E.Y., Li, S.Y., Wang, L.X., Zhang, C.G., Zhao, Y.H., 2015. Revalidation and re-description of Brachymystax tsinlingensis Li, 1966 (Salmoniformes: Salmonidae) from China. Zootaxa, 3962: 191-205. https://doi.org/10.11646/zootaxa.3962.1.12
Yu, J.N. and Kwak, M., 2015. The complete mitochondrial genome of Brachymystax lenok tsinlingensis (Salmoninae: Salmonidae) and its intraspecific variation. Gene, 573: 246-253. https://doi.org/10.1016/j.gene.2015.07.049










