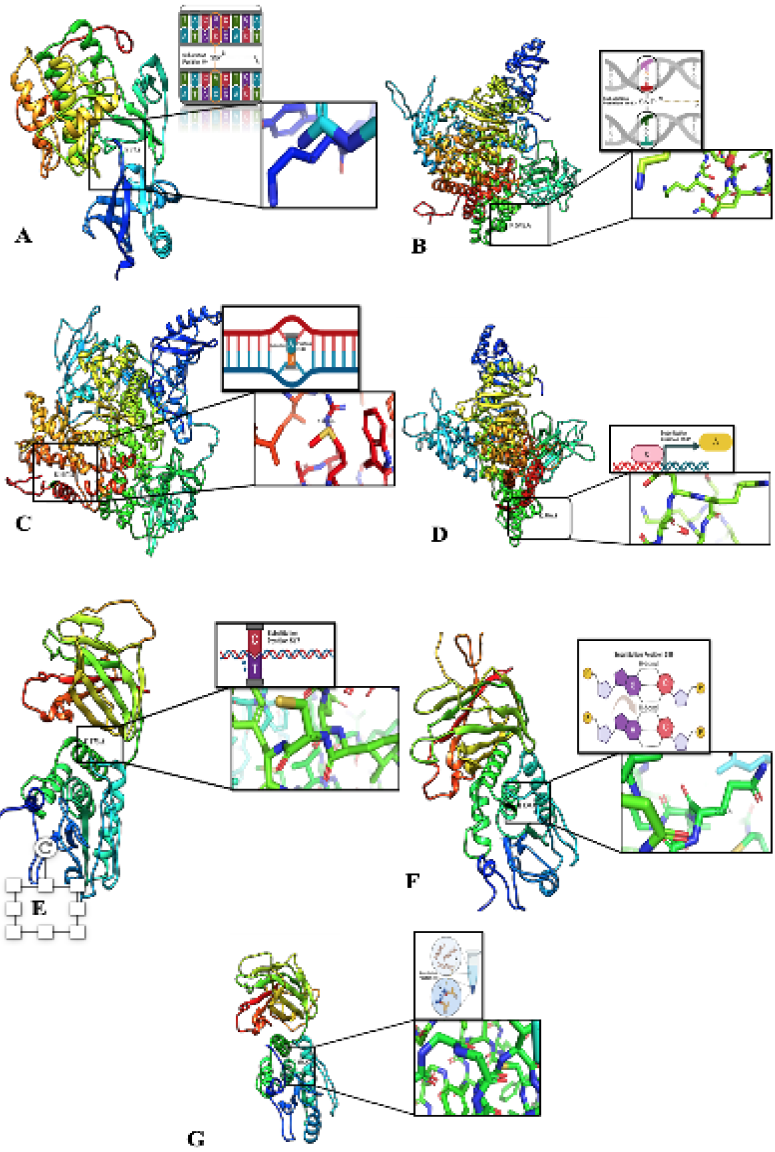
3D-modelled structure from SNPs sequence. A, Structural modeling reveals AKT1 gene mutation (E17K) on Chromosome 14, Exon 3, position 49 (c.49G>A) causing a missense mutation Amino acids change (E to K) with potential clinical association. B, An In-depth Analysis of the Structural Implications of the c.166G>A PIK3CA Gene Mutation (E545K) in Exon 10 on Chromosome 3, Resulting in a Missense Mutation that Substitutes Glutamic Acid (E) with Lysine (K) in the PIK3CA Protein. C, Protein Modeling of PIK3CA Gene Mutation (H1047R) in Exon 21 (c.31406A>G) on Chromosome 3: Revealing Molecular Mechanisms of a Missense Mutation Substituting Histidine with Arginine. D, PIK3CA Gene Mutation: A Missense Q546K Change in Exon 10 (c.1636 C>A) - A Promising Protein Model Insight. E, Modelling depict PTEN gene mutation P.R 173c (chromosome 10, Exon 6). c.517 C>T results in a missense mutation, substituting Arginine (R) with Cysteine (C) at position 517, offering insights into its potential impact on PTEN protein structure and function. F, In silicon protein modeling predicts the impact of PTEN gene mutation (P.R130Q) on chromosome 10, Exon 5, c.389G>A substitution, causing a missense mutation (R to Q). G, Mutation profiling and protein modeling to predict the location and effects of the PTEN gene mutation P.R130G on chromosome 10, exon 5 this missense mutation (c. C>G, position 388) results in an amino acid change from arginine (R) to cysteine (C).”