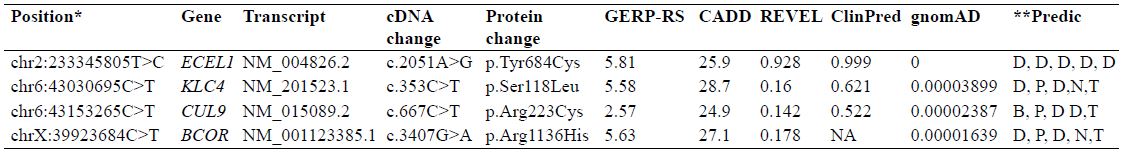
Table II.
Segregating variants in family RDHR-01.
*, position according to the human genome build GRCh37/hg19; GERP-RS, genomic evolutionary rate profiling-rejected substitutions; CADD, combined annotation dependent depletion; REVEL, rare exome variant ensemble learner; ClinPred, (https://sites.google.com/site/clinpred/); ExAC, exome aggregation consortium; ,**, predictions according to PolyPhen2; MutationTaster, PROVEAN, SIFT and FATHMM, respectively; D, deleterious or damaging, P, pathogenic; N, neutral; T, tolerated; B, benign; NA, not available.