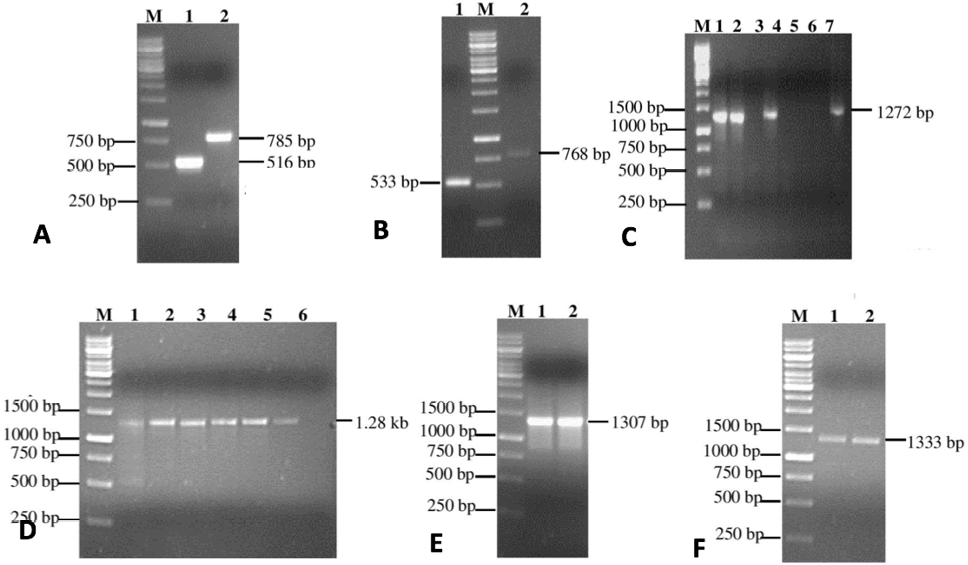
Analysis of PCR products by 1% agarose gel electrophoresis. Gels were visualized by staining with ethidium bromide; A, Gel purified PCR products of steps # 1 of SOE-PCR. Lane M: DNA size marker, Lane 1: IFNα2b gene (516 bp) amplified using primer pair F1 and F221, Lane 2: LAP gene (785 bp) amplified using primer pair F3 and F4; B, Gel purified PCR products of steps # 1 of SOE-PCR . Lane M: DNA size marker, Lane 1: IFNα2b gene (533 bp) amplified using primer pair F3/ and F4/, Lane 2: LAP gene (768 bp) amplified using primer pair F1/ and F2/21; C, Optimization of annealing temperature (44-60 ͦ C) in first 10 cycles of step # 2 of SOE-PCR during which 30 bp overhang (HCV NS3 protease cleavage site sequence ) at the 3’ end of IFNα2b gene (516 bp) overlap by 21 bp with 5’ end overhang of LAP gene (774 bp). In later 30 cycles, 58 °C annealing temperature was used with primer pair F1 and F4 to get 1.28 kb chimeric gene product. Lane M: DNA size marker, Annealing temperatures in Lane 1: 50 ͦ C, Lane 2: 55 ͦ C, Lane 3: 60 ͦC, Lane 4: 58 ͦ C, Lane 5: 48 ͦ C, Lane 6: 44 ͦ C and in Lane 7: 58 ͦ C; D, Optimization of minimum required equimolar concentration of genes for SOE-PCR. Lane M: DNA size marker, PCR product using equimolar concentration in Lane 1: 0.80 pmoles, Lane 2: 0.20 pmoles, Lane 3: 0.10 pmoles, Lane 4: 0.05 pmoles, Lane 5: 0.025 pmoles, Lane 6: 0.0125 pmoles and Lane 7: 0.00625 pmoles; E, 1st step of OPW-PCR with F5/F4 and F5//F4/ primer using chimeric genes IFNα2b-NS3-LAP and LAP-NS3-IFNα2b respectively introduced enterikinase cleavage site and Gly-Ser spacer at 5’ end; F, 2nd round of OPW-PCR with F6/F4 and F6/F4/ primer using PCR products of 1st round of OPW-PCR as templates to introduce His-Tag sequence and XhoI cleavage site.