Molecular Characterization of Plasmodium relictum in Four Common Bird Species in India
Molecular Characterization of Plasmodium relictum in Four Common Bird Species in India
Gautam Patra1*, Ana Sahara2, Sonjoy Kumar Borthakur1, Parthasarathi Behera3, Subhamoy Ghosh1, Apurba Debbarma4 and Seikh Sahanawaz Alam5
1Department of Veterinary Parasitology, Central Agricultural University, India
2Department of Veterinary Parasitology, Gadjah Mada University, Yogyakarta, Indonesia
3Department of Veterinary Physiology and Biochemistry, College of Veterinary Sciences and Animal Husbandry, Central Agricultural University, Selesih, Aizawl, Mizoram, India.
4Department of Veterinary Parasitology, College of Veterinary Sciences and Animal Husbandry, R.K. Nagar, Agartala, Tripura, India
5Department of Botany, Garhbeta College, Paschim Midnapore, West Bengal, India
ABSTRACT
The increasing emergence of wildlife diseases with the possibility to ecological threats as well as domestic animals and human health has prompted the importance of understanding disease dynamics and associated risks in biological conservation. The present study was undertaken from North Eastern part of India from January, 2017 to March, 2018 to identify and molecularly characterize Plasmodium relictum based on cyt b gene in various species of wild birds (Upupa epops, Passer domesticus, Pycnonotus cafer, Bubulcus ibis). The birds were captured by netting system. After blood was collected from wing veins, birds were released from the cages. Blood samples were examined after staining with Giemsa stain. The positive samples were used for amplification of cyt b gene of Plasmodium relictum. Cloning and sequencing of the amplified products of each samples was performed separately. Out of 120 birds examined, 25 were found positive for P. relictum based on morphology and subsequently confirmed by PCR which selectively amplified 712bp of P. relictum. Based on sequences and phylogenetic analysis of cyt b gene, 8 isolates of P. relictum were identified. The obtained complete nucleotide sequences of cyt b gene from P. relicum revealed100% identity among themselves while the other sequences registered in Gen Bank showed 95% to 99% similarity. In the phylogenetic analysis all the isolates of P. relictum formed a separate clade with high bootstrap values. It can be inferred from the study that P. relictum is fairly common in wild birds and the cyt b gene is highly conserved among different isolates. It seems that cytb could provide suitable genetic markers for discrimination and genetic characterization of P. relictum. This is the first attempt of genetic characterization of P. relictum from India.
Article Information
Received 22 September 2018
Revised 01 March 2019
Accepted 03 May 2019
Available online 24 December 2020
Authors’ Contribution
PB, SG, AD and SSA collected the blood samples. Parasite identification and molecular analysis was done by GP and AS. GP and SKB critically supervised the whole experiment and drafted the manuscript.
Key words
Plasmodium relictum, cyt b gene, Molecular characterization, India
DOI: https://dx.doi.org/10.17582/journal.pjz/20180922080933
* Corresponding author: dr.gautampatra@yahoo.co.in
0030-9923/2021/0001-0305 $ 9.00/0
Copyright 2021 Zoological Society of Pakistan
INTRODUCTION
Plasmodium relictum infects a wide variety of domestic and wild birds including chickens, ducks, patridges, canaries, pigeons etc (Bensch et al., 2009; Marzal et al., 2014; Drovetski et al., 2014). It may be lethal to the species which have not evolved resistance to parasites such as penguins and naïve birds which often suffer from severe disease and mortality during infection (Ilgūnas et al., 2016). The parasites are easily distinguishable in blood films because of its distinct morphological characteristics. Mature stages typically have predominant nuclei and cytoplasm, numerous pigment granules and markedly influence the position of host cell nuclei causing lateral shifts in their positions. Microscopic examination of blood films was main diagnostic tool in 20th century worldwide (Atkinson, 2008). Microscopy may reveal the presence of trophozoites, merozoites and gametocytes in erythrocytes.
Several molecular methods are used for detecting avian malarial parasites in blood samples. Most are based on amplification of conserved fragments of 18S rRNA or mitochondrial cytochrome-b gene (Jarvi et al., 2002; Tattiyapong et al., 2016; Valkiūnas et al., 2018). Partial sequences of cyt b gene have been successfully used for molecular characterization of parasites and it proved to be excellent molecular marker for disease diagnostics (Elsasser et al., 2009).
Despite wide spread distribution of P. relictumin birds, no study has been done in India to characterize the cyt b gene and also about the number of copies it possesses. Therefore, the objective of this study is to identify comprehensive morphological features of blood stages of the parasites and the genetic diversity of the targeted gene. This will give new directions for future avian malaria research.
MATERIALS AND METHODS
Collection of blood samples
Field work was carried from various states of North Eastern India from January, 2017 to March 2018. Sixty three common hoopoes (Upupaepops), twenty seven house sparrows (Passer domesticus), twenty two red vented bulbuls (Pycnonotus cafer) and eight egrets (Bubulcus ibis) were caught with mist nets and large stationary traps. The blood samples were taken by puncturing the wing vein. Blood films were prepared immediately, air dried, fixed in methanol and stained with Giemsa’s stain following standard procedure (Soulsby, 1982). About 50µl of whole blood was taken in EDTA coated vials and stored at ambient temperature in the field and then stored at -20°C in the laboratory until further use.
Isolation of genomic DNA from blood samples
Microscopically positive blood samples (25) for Plasmodium relictum were subjected to genomic DNA isolation. DNA was isolated from 10µl of EDTA anticoagulated whole blood of 25 positive samples. Twenty hoopoe, two from house sparrow, two from red vented bulbul and one from egret using DNeasy Blood and Tissue Kit (Qiagen, Germany) following the manufacturer’s protocol and stored at -20°C for downstream use.
PCR amplification and purification
The Plasmodium cyt b gene specific primer set (PlasF: GAGAATTATGGAGTGGATGGTG and PlasR: GTGGTAATTGACATCCWATCC) was used in this study to amplify the partial sequence of cyt b gene with minor modifications. The PCR reaction was carried out in 25µl of 1X PCR green buffer (Thermo Scientific, USA) containing 0.2 µl of Dream Taq DNA polymerase, 10 pmol of each primer and 0.2 mM concentration of each deoxyribonulceotidet riphsophate, with 3µl of template DNA. Amplification was performed using a C1000 thermal cycler (BioRad, USA) under following conditions: initial denaturation at 94°C for 5 min, followed by 35 amplification cycles (94°C for 30 sec, 55°C for 45 sec and 72°C for 1 min) and a final extension at 72°C for 10 min. Nested PCR was done to amplify 712 bp gene fragment of cyt b region following the same thermal cycling conditions. The PCR amplicons were checked by electrophoresis on 1.5% low melting agarose gel and purified by gel extraction by using Qiaex II gel extraction kit (Qiagen, Germany) following manufacturer’s protocol.
Cloning and sequencing
For cloning of cyt b gene, the purified PCR products from each sample were separately ligated into pTZ57R/T TA cloning vector (Thermo Scientific, USA) and incubated at 4°C for overnight. The plasmid DNA constructs were transformed into competent DH5α Escherichia coli cells using Ins TAclone PCR cloning kit (Thermo Scientific, USA) according to manufacturer’s protocol. The transformed cells were plated immediately on pre-warmed LB agar plates supplemented with ampicillin (100µg/ml), X-gal (30µg/ml) and IPTG (0.5mM/ml) for the development of blue white colonies. The positive clones were confirmed by colony PCR using specific primers. The stab cultures of two positive clones per sample containing the desired gene were custom sequenced from the Department of Biochemistry, Delhi University, South Campus, India. The fragments were sequenced at least twice to reduce possibility of sequencing errors.
Sequence analysis
All the newly generated sequences of cyt b gene of Plasmodium relictum isolates were compared with each other and with published sequences in the nucleotide database in Gen Bank using the BLAST programme of the National Centre for Biotechnology Information and aligned by Clustal W.
Phylogenetic analysis
Phylogenetic analysis of P. relictum of the eight isolates collected from different geographical areas of NE region of India were done independently with the sequences of cyt b genes. A total of twenty gene sequences including eight newly generated sequences were used for analysis. Phylogenetic analysis was done by using the MEGA6.0 software (Tamura et al., 2013) with the maximum likelihood method. The nucleotide substitution model for best fit to the data set was evaluated in MEGA6.0. Tamura-Nei model (TN93) with gamma distribution (TN93+G) was found to be the model of choice for phylogenetic analysis of cyt b gene. The phylogeny was analysed using 1000 bootstrap replications. Plasmodium falciparum species from human was used as out group species to root the tree.
Prevalence study
The prevalence of Plasmodium relictum was recorded according to age and species of birds. The prevalence (P) was estimated according to standard method (Thrusfield, 2007) and by the formula as given below:

Statistical analysis
The upper and lower limits of the confidence intervals for the infection values in each species were calculated by the following formula (Bawm et al., 2016)
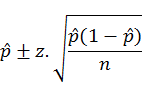
where, P-hat is dividing the numbers of events by the number of trials; z-score is the table value and n is the number of samples
Ethical statement
All animal experiments were carried out strictly as per the guidelines issued by ARRIVE and Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) and was approved by Institutional Animal Ethics Committee with reference No. CVSc/CAU/IAEC/no. 6641, dtd, Selesih, the 25th, April, 2016.
RESULTS
Parasitic identification and prevalence
Microscopic examination of the Giemsa stained thin smear revealed that 25 (21%) out of 120 blood samples were positive for Plasmodium relictum (Table I). The trophozoite, schizont stages were observed in the cytoplasm of the red blood cells (Fig. 1C). From morphological characteristics, the parasites were identified as P. relictum. P. relictum can be distinguished from other plasmodial parasites of birds by the presence of round or irregular gamonts inside RBC with fine and pinpoint pigment granules. Out of 63 hoopoes 20 (16.66%) were found positive for P. relictum, 02 (1.66%) of 27 blood samples of house sparrow, 02 (1.66%) of 22 red vented bulbuls and 01(0.83) of 08 egrets also showed positive (Table II). The prevalence of P. relictum was found higher in adult (15.83%) birds than sub-adults (5%).
PCR amplification and cloning of cyt b gene
The microscopic finding of Plasmodium spp. in red blood cells was confirmed by PCR. The amplified products of cyt b gene were checked by ethidium bromide stained agarose gel (1.5%) electrophoresis which showed ~ 712 bp size of the desired gene. The PCR products were purified, cloned into pTZ57R/T TA cloning vector and confirmed by colony PCR (Fig. 2). Subsequently all isolates were custom sequenced.
Sequence analysis of newly generated cyt b region
Eight sequences of the cyt b gene (712bp) generated in this study were submitted to GenBank (Table II). Sequence similarity searches in BLAST showed all isolates of the targeted genes have high similarity with USA and Germany isolates (Fig. 3). A closer comparison of the newly generated 8 isolates had 100% identity among themselves and 99.6% with USA (DQ659566) and 99.3% with Germany (MF189958) isolates, respectively. These 8 new sequences did not show any difference in nucleotide position.
Table I. Overall prevalence rate of Plasmodium relictum in birds.
|
Total number of birds examined |
Number of birds infected (%) |
Range |
|
|
120 |
25 (21) |
||
|
Hoope |
20 (16.66) |
12.07-21.26 |
|
|
House sparrow |
02 (1.66) |
0.09-3.24 |
|
|
Red vented bulbul |
02 (1.66) |
0.09-3.24 |
|
|
Egret |
01 (0.83) |
0.29-1.95 |
|
|
Young |
|||
|
Sub adult |
06 (5) |
2.31-7.69 |
|
|
Adult |
19 (15.83) |
11.32-20.33 |
|
Phylogenetic relationship
The phylogenetic tree based on maximum likelihood method with Tamura-Nei model for distance calculation based on the 712 bp region of cyt b gene is shown in Figure 4. P. relictumfrom those areas were compared with P. gallinaceum and P. falciparum. Pasmodium relictum in this research was located in one clade which showed that they are genetically closer to Plasmodium gallinaceum than to P. falciparum. Isolates from Spain (KT363870), Sweden
Table II. Species of Plasmodium relictum from which cyt b sequences were used for phylogenetic analysis together with their location/host and GenBank accession number.
|
Species |
Region of origin |
Host |
GenBank accession no. |
|
P. relictum |
Mizoram |
Hoopoe (Upupa epops) |
MH373249* |
|
P. relictum |
Mizoram |
House sparrow (Passer domesticus) |
MH373250* |
|
P. relictum |
Manipur |
Red vented bulbul (Pycnonotus cafer) |
MH373251* |
|
P. relictum |
Manipur |
Cattle egret (Bubulcus ibis) |
MH373252* |
|
P. relictum |
Tripura |
Cattle egret (Bubulcus ibis) |
MH373253* |
|
P. relictum |
Tripura |
Red vented bulbul (Pycnonotus cafer) |
MH373254* |
|
P. relictum |
Nagaland |
Hoopoe (Upupa epops) |
MH373255* |
|
P. relictum |
Nagaland |
House sparrow (Passer domesticus) |
MH373256* |
|
Plasmodium sp. |
USA |
Red-billed quelea (Quelea quelea) |
DQ659566 |
|
P. relictum |
Japan |
Culexfuscanus |
AB308046 |
|
P. relictum |
Lithuania |
Common canary (Serinus canaria), European goldfinch (Carduelis carduelis), Zebra finch (Taeniopygia guttata), Budgerigar (Melopsittacus undulatus) |
MG724747 |
|
P. relictum |
Sweden |
Sudan golden sparrow (Passer luteus) |
AF495571 |
|
P. relictum |
Sweden |
Eurasian blackcap (Sylvia atricapilla) |
AY831748 |
|
P. relictum |
Sweden |
Great reed warbler (Acrocephalus arundinaceus) |
AF254975 |
|
P. relictum |
USA |
African penguin (Spheniscus demersus) |
KY653774 |
|
P. relictum |
Germany |
Carrion crow (Corvus corone) |
MF189958 |
|
Plasmodium sp. |
USA |
Tufted titmouse (Baeolophus bicolour) |
GQ141593 |
|
P. relictum |
Spain |
House sparrow (Passer domesticus) |
KT363870 |
|
P. gallinaeceum |
USA |
Red jungle fowl (Gallus gallus) |
AY099029 |
|
P. falciparum |
New Delhi, India |
Homo sapiens |
AJ298787 |
*New sequences generated in this present study.
(AF254975) formed separate clade in the neighbour joining tree with high bootstrap support (86%) by removing P. falciparum (AJ298787) from the analysis as out-group species. Isolates from Lithuania (MG724747) and Sweden (AY83178) was found distantly related with the present isolates. Another neighbour joining tree with the different P. relictum isolates from the present investigation was constructed to check the intra-species variation in relation to each other, if any (Fig. 5).
DISCUSSION
On the basis of morphology, the parasites were identified as Plasmodium relictum. However, the microscopic identification is not sufficient to differentiate different species of Plasmodium of birds. It requires extensive skills and experience, especially during early and chronic stages of infection (Silveira et al., 2009; Silveira et al., 2013). In the present study, all four species of wild birds were found infected with P. relictum with varying intensity. Several studies have revealed a lack of host specificity in Plasmodium infection (Zhang et al., 2014; Silva-Iturriza et al., 2012; Beadell et al., 2009; Waldenström et al., 2002). This means that host switching between avian species is more likely to occur in Plasmodium. Adult birds were found more infected than sub-adult groups, probably because of the time between an infection being transmitted to a nestling and parasites entering the nestling stage (Valkiūnas, 2005). After infections, individuals either become ill or die during the active phase. Alternatively, they may develop an effective immune response and clear the infection. As a result, older birds become less susceptible as they grow old. On the other hand, an age dependent deterioration of the immune system lead to a late-life increase in infection compared to sub-adult groups (Palacios et al., 2007; Palacios et al., 2011). No other haemoparasites were detected in the four selected species of birds. However, we only sampled from NE states of India, so additional studies are needed from other locations of India to determine haemoprotozoan diversity.
The microscopic findings of Plasmodium relictum in RBCs of four species of wild birds was confirmed by PCR. PCR was found very sensitive to detect the infection in birds. This result was similar to the study reported by Tattiyapong et al. (2016) and Krams et al. (2012) who also reported sensitivity of PCR during the detection of haemoparasites infection.
The 712bp PCR amplicon from all positive samples were cloned and subsequently sequenced. The sequences of 8 samples showed clear sequencing patterns. These sequence data were then deposited in NCBI Gen Bank database under accession numbers: MH373249-MH373256. Sequence similarity between the partial cyt b gene fragments of all isolates was 100% with no nucleotide substitution. The multiple nucleotide sequence alignments of P. relictum showed that the cyt b gene in the study region was highly conserved.
On the basis of sequence homology and phylogenetic analysis of cyt b gene all isolates of Plasmodium sp. were confirmed. The phylogenetic analysis revealed that all P. relictum isolates were clustered in the same clade and separated from P. falciparum. Similar studies have been reported from different geographical locations (Tattiyapong et al., 2016; Silveira et al., 2013; Murata et al., 2008; Chen et al., 2015). No intra-specific variation was observed among different isolates collected from North-East India suggesting that all isolates were identical. To date, mitochondrial fragments of cyt b gene has not been characterized from Plasmodium sp. of birds from India. Here we have presented the partial sequence of cyt b gene of Plasmodium relictum. Neighbour joining tree shows strong support for Plasmodium and apicomplexan clade (95% bootstrap support).
In conclusion, we morphologically and genetically characterized P. relictum from four species of wild birds in NE region of India. The present study demonstrates that P. relictum is fairly common in wild birds and has the conserved gene sequence encoding for cytochrome oxidase b. Along with the microscopic examination, this report first describes the molecular characterization of P. relictum of wild birds from NE region of India. It also highlights the importance of isolating and characterizing other Plasmodium mitochondrial genes which would provide additional information into codon usage and insights about the evolution of apicomplexan parasites. However, further studies with larger number of samples from various wild birds as well as vectors involved in different geographical areas covering other parts of India should be carried out for determining the status and the genetic diversity of avian malaria parasites in India.
ACKNOWLEDGEMENTS
The authors duly acknowledged the Dean, College of Veterinary Sciences and A.H., Central Agricultural University, Selesih, Aizawl, Mizoram for providing necessary facilities to conduct the study.
Statement of conflict of interest
The authors declare that there is no conflict of interest
Funding source
The present study was funded by the Department of Veterinary Parasitology, CVSc and AH, CAU for post graduate research work under the supervision of first author (No.CAU/CVSc/PG Res/Para/2017).
REFERENCES
Atkinson, C.T., 2008. Avian malaria. In: Parasitic diseases of wild birds (eds. C.T. Atkinson, N.J. Thomas and D.B. Hunter), Wiley-Blackwell Publishing, Iowa. pp. 35-53. https://doi.org/10.1002/9780813804620.ch3
Bawm, S., Htun, L.L., Maw, N.N., Ngwe, T., Tosa, Y., Kon, T. and Katakura, K., 2016. Molecular survey of Babesia infections in cattle from different areas of Myanmar. Ticks Tick-borne Dis., 7: 204-207. https://doi.org/10.1016/j.ttbdis.2015.10.010
Beadell, J.S., Covas, R., Gebhard, C., Ishtiaq, F., Melo, M., Schmidt, B.K. and Fleischer, R.C., 2009. Host associations and evolutionary relationships of avian blood parasites from West Africa. Int. J. Parasitol., 39: 257–266. https://doi.org/10.1016/j.ijpara.2008.06.005
Bensch, S., Hellgren, O. and Pérez-Tris, J. 2009. MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Res., 9: 1353-1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x
Chen, T.H., Aure, W.E., Cruz, E.I., Malbas, F.F.Jr., Teng, H.J., Lu, L.C., Kim, K.S., Tsuda, Y. and Shu, P.Y., 2015. Avian Plasmodium infection in field-collected mosquitoes during 2012-2013 in Tarlac, Philippines. J. Vector Ecol., 40: 386-392. https://doi.org/10.1111/jvec.12178
Drovetski, S.V., Aghayan, S.A., Mata, V.A., Lopes, R.J., Mode, N.A. and Harvey, J.A., 2014. Does the niche breadth or trade-of hypothesis explain the abundance—occupancy relationship in avian Haemosporidia? Mol. Ecol., 23: 3322–3329. https://doi.org/10.1111/mec.12744
Elsasser, S.C., Floyd, R., Hebert, P.D.N. and Schulte-Hostedde, A.I., 2009. Species identification of North American guinea worms (Nematoda: Dracunculus) with DNA barcoding. Mol. Ecol. Res., 9: 707–712. https://doi.org/10.1111/j.1755-0998.2008.02393.x
Ilgūnas, M., Bukauskaitė, D., Palinauskas, V., Iezhova, T.A., Dinhopl, N. and Nedorost, N., 2016. Mortality and pathology in birds due to Plasmodium (Giovannolaia) homocircumfexum infection, with emphasis on the exoerythrocytic development of avian malaria parasites. Malaria J., 15: 256. https://doi.org/10.1186/s12936-016-1310-x
Jarvi, S.I., Schultz, J.J. and Atkinson, C.T., 2002. PCR diagnostics underestimate the prevalence of avian malaria (Plasmodium relictum) in experimentally-infected passerines. J. Parasitol., 88: 153-158. https://doi.org/10.1645/0022-3395(2002)088[0153:PDUTPO]2.0.CO;2
Krams, I., Suraka, V., Cirule, D., Hukkanen, M., Tummeleht, L., Mierauskas, P., Rytkönen, S., Rantala, M.J., Vrublevska, J., Orell, M. and Krama, T., 2012. A Comparison of microscopy and PCR diagnostics for low intensity infections of haemosporidian parasites in the Siberian Tit Poecilec inctus. Annls. Zool. Fenn., 49: 331-340. https://doi.org/10.5735/086.049.0506
Marzal, A., García-Longoria, L.J.M., Cárdenas, C. and Sehgal, R.N., 2015. Invasive avian malaria as an emerging parasitic disease in native birds of Peru. Biol. Invasions, 17: 39–45. https://doi.org/10.1007/s10530-014-0718-x
Murata, K., Nii, R., Sasaki, E., Ishikawa, S., Sato, Y., Sawabe, K., Tsuda, Y., Matsumoto, R., Suda, A. and Ueda, M., 2008. Plasmodium (Bennettinia) juxtanucleare infection in acaptive white eared-pheasant (Crossoptilon crossoptilon) at a Japanese zoo. J. Vet. med. Sci., 70: 203-205. https://doi.org/10.1292/jvms.70.203
Palacios, M.G., Cunnick, J.E., Winkle,r D.W. and Vleck, C.M., 2007. Immuno senescence in some but not all immune components in a free-living vertebrate, the tree swallow. Proc. R. Soc. B., 274: 951–957. https://doi.org/10.1098/rspb.2006.0192
Palacios, M.G., Winkler, D.W., Klasing, K.C., Hasselquist, D. and Vleck, C.M., 2011. Consequences of immune system aging in nature: a study of immunosenescence costs in free-living tree swallows. Ecology, 92:952–966. https://doi.org/10.1890/10-0662.1
Silva-Iturriza, A., Ketmaier, V. and Tiedemann, R., 2012. Prevalence of avian haemosporidian parasites and their host fidelity in the central Philippine islands. Parasitol. Int., 61: 650–657. https://doi.org/10.1016/j.parint.2012.07.003
Silveira, P., Damatta, R.A. and Dagosto, M., 2009. Hematological changes of chicken sex perimentally infected with Plasmodium (Bennettinia) juxtanucleare. Vet. Parasitol., 162: 257-262. https://doi.org/10.1016/j.vetpar.2009.03.013
Silveira, P., Marin, S.Y., Moreira, P.A., Tocantins, B.B., Lacorte, G., Paixao, T.A., Martins, N.R. and Braga, E.M., 2013. Interactions of Plasmodium juxtanucleare and chicken anaemia virus: establishing a model. Parasitology, 140: 1777-1788. https://doi.org/10.1017/S0031182013001170
Soulsby, E.J.L., 1982. Helminths, arthropods and protozoa of domesticated animals. Baillere Tindall Publication. 1982.
Tamura, K., Stecher, G., Peterson, D., Filipski, A. and Kumar, S., 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol., 30: 2725-2729. https://doi.org/10.1093/molbev/mst197
Tattiyapong, M., Deemagarn, T., Mohkeaw, K., Ngamjiteu, S. and Jiratanh, M., 2016. Molecular characterization of Plasmodium juxtanucleare in Burmese red jungle fowls (Gallus gallusspadiceus) in Thailand. J. Protozool. Res., 26: 1-10.
Thrusfield, M., 2007. Veterinary epidemology. Chapter 5 In: Determinants of disease. Third edition. Blackwell publishing. pp.76.
Valkiūnas, G., 2005. Avian malaria parasites and other haemosporidia. CRC press. https://doi.org/10.1201/9780203643792
Valkiūnas, G., Ilgūnas, M., Bukauskaitė, D., Fragner, K., Weissenböck, H., Atkinson, C.T. and Iezhova, T.A., 2018. Characterization of Plasmodium relictum, a cosmopolitan agent of avian malaria. Malaria J., 17: 184. https://doi.org/10.1186/s12936-018-2325-2
Waldenström, J., Bensch, S., Kiboi, S., Hasselquist, D., Ottosson, U., 2002. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol. Ecol., 11: 1545–1554. https://doi.org/10.1046/j.1365-294X.2002.01523.x
Zhang, Y., Wu, Y., Zhang, Q., Su, D., Zou, F., 2014. Prevalence Patterns of avian Plasmodium and Haemoproteus parasites and the influence of host relative abundance in Southern China. PLoS One, 9: e99501. https://doi.org/10.1371/journal.pone.0099501
To share on other social networks, click on any share button. What are these?









