Identification and Expression Patterns of Chemosensory Genes in Male and Female Wax Moths, Galleria mellonella
Identification and Expression Patterns of Chemosensory Genes in Male and Female Wax Moths, Galleria mellonella
Shuang Yang1,2, Huiting Zhao3, Xuewen Zhang2, Kai Xu1, Lina Guo1, Yali Du1 and Yusuo Jiang1,*
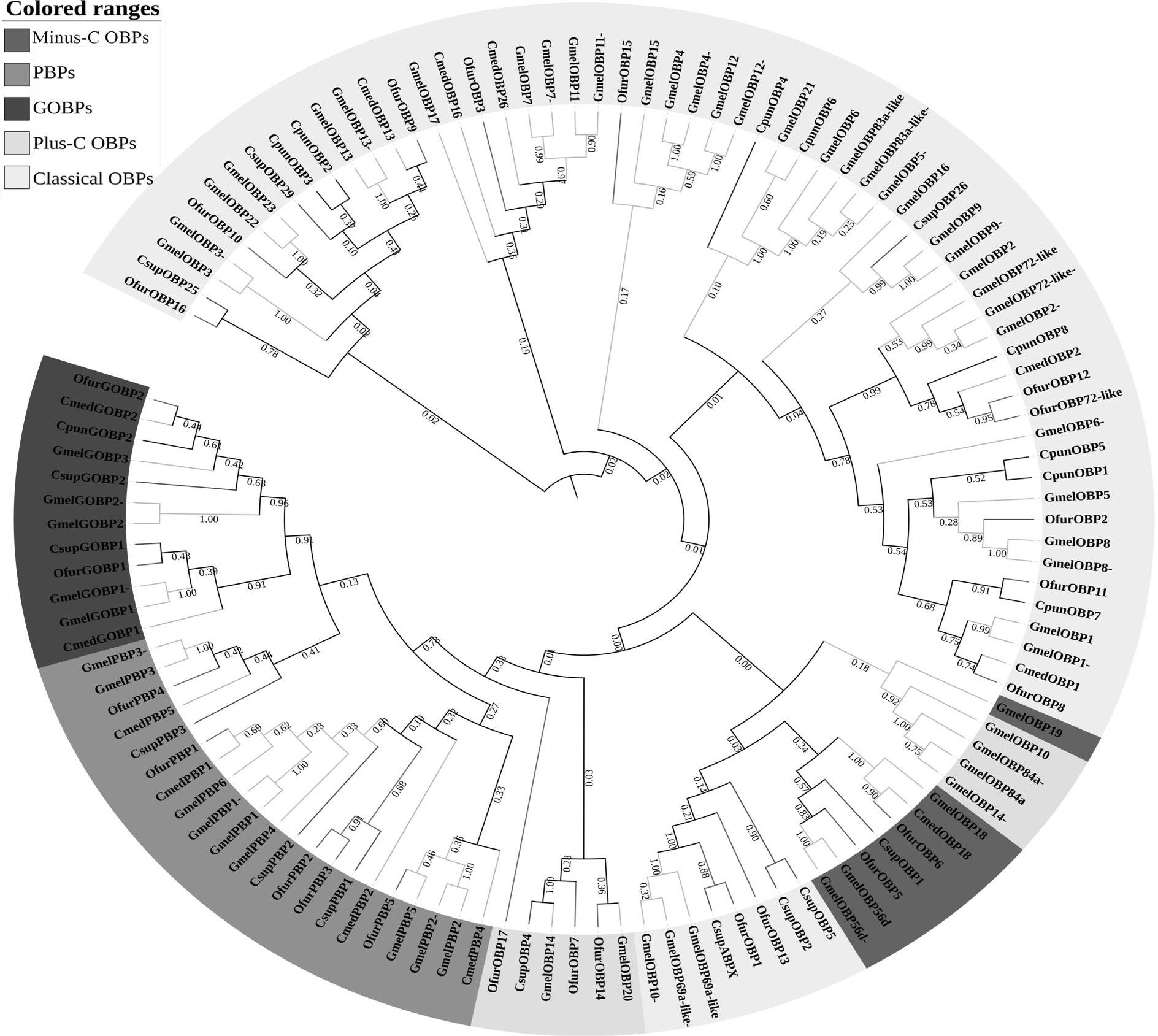
Phylogenetic tree of candidate GmelOBPs with known lepidopteran odorant-binding protein (OBP) sequences. Gmel, Galleria mellonella; Csup, Chilo suppressalis; Ofur, Ostrinia furnacalis; Cmed, Cnaphalocrocis medinalis; Cpun, Conogethes punctiferalis. The clade in green and purple indicates the PBPs and GOBPs, respectively; the Minus-C OBPs subfamily is marked in red and the Plus-C OBPs subfamily is marked in blue; the Classical OBPs subfamily is marked in yellow.
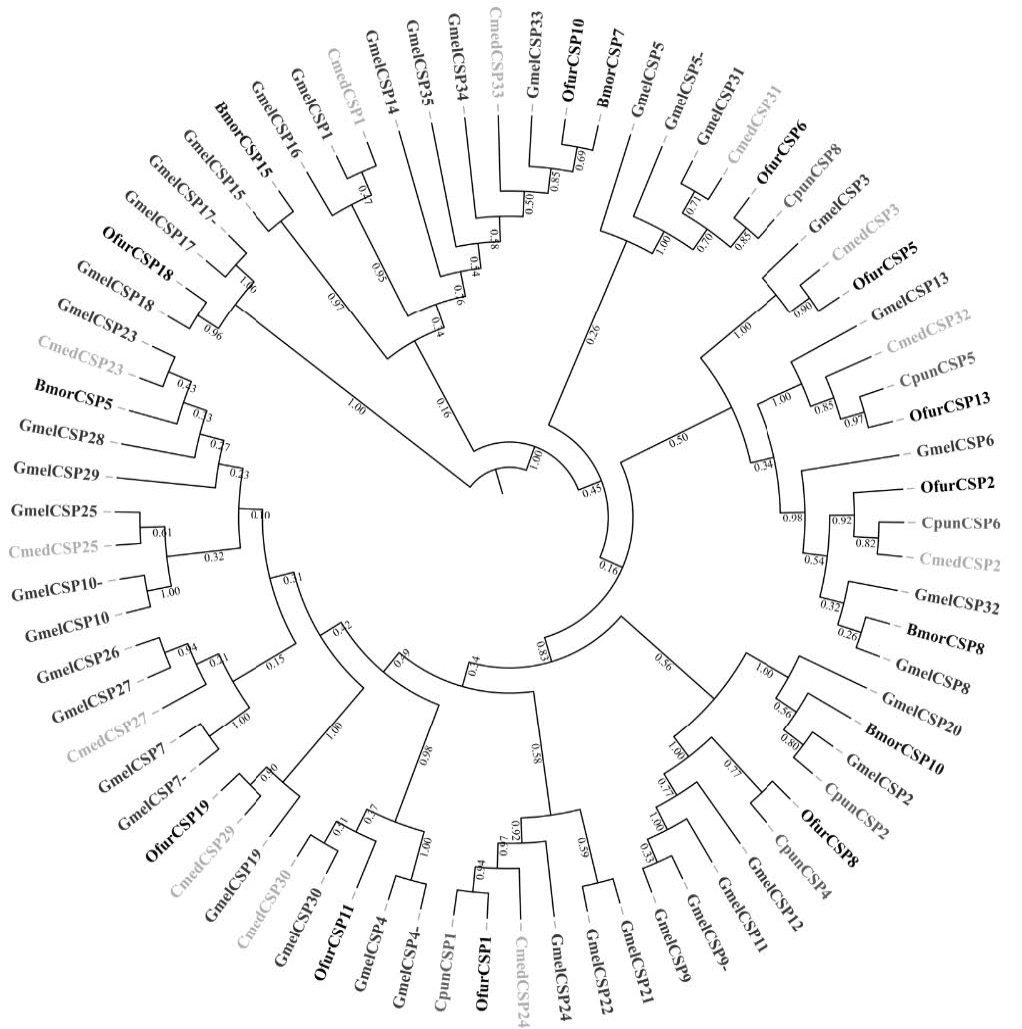
Phylogenetic tree of candidate GmelCSPs with known lepidopteran chemosensory protein (CSP) sequences. Gmel, Galleria mellonella; Csup, Chilo suppressalis; Ofur, Ostrinia furnacalis; Cmed, Cnaphalocrocis medinalis; Cpun, Conogethes punctiferalis; Bmor, Bombyx mori.
Phylogenetic tree of candidate GmelORs with known lepidopteran olfactory receptor (OR) sequences. Gmel, Galleria mellonella; Csup, Chilo suppressalis; Ofur, Ostrinia furnacalis; Cmed, Cnaphalocrocis medinalis; Cpun, Conogethes punctiferalis; Bmor, Bombyx mori. The clade in purple indicates the Orco receptor gene clade.
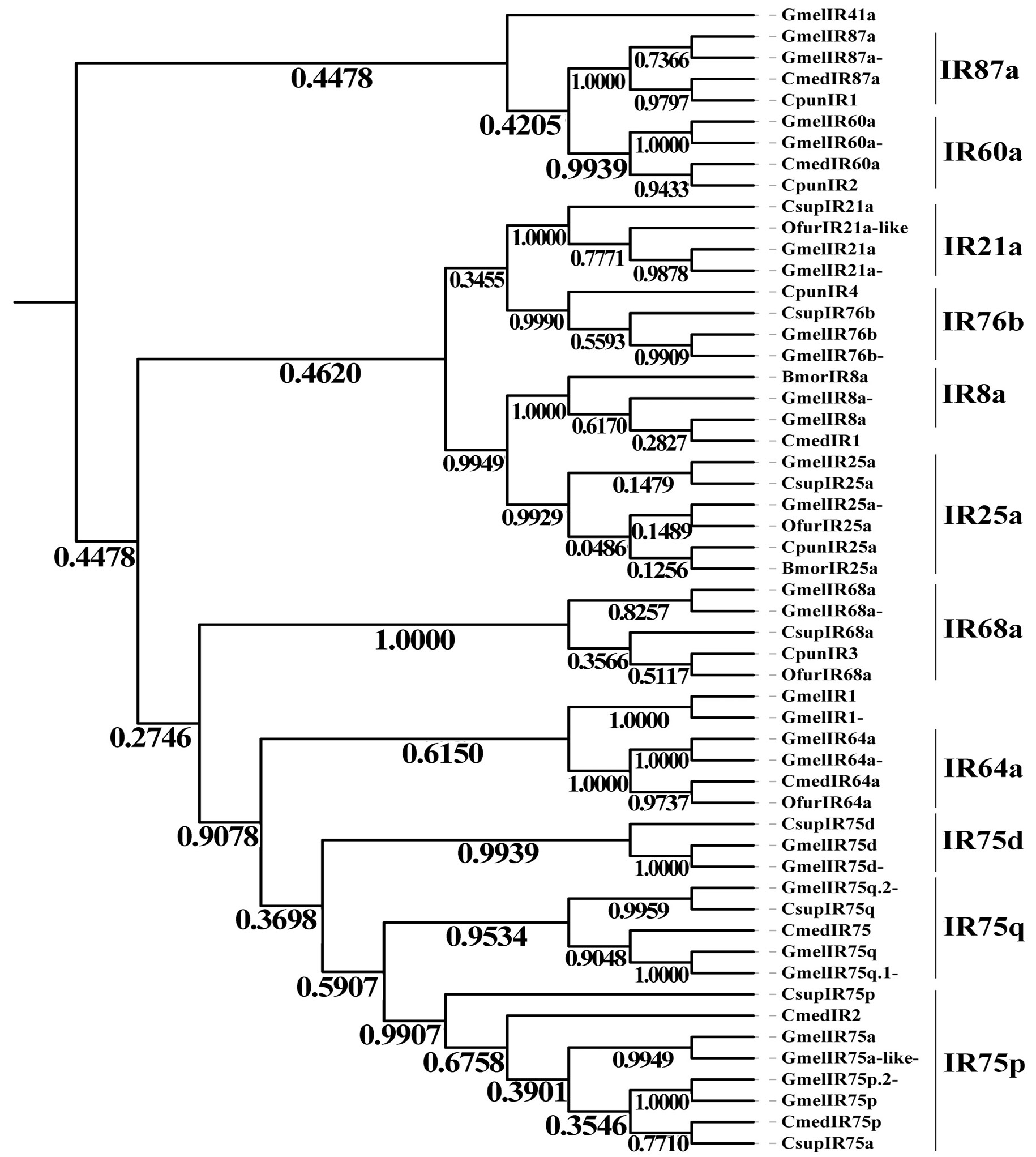
Phylogenetic tree of candidate GmelIRs with known lepidopteran ionotropic receptor (IR) sequences. Gmel, Galleria mellonella; Csup, Chilo suppressalis; Ofur, Ostrinia furnacalis; Cmed, Cnaphalocrocis medinalis; Bmor, Bombyx mori; Cpun, Conogethes punctiferalis.
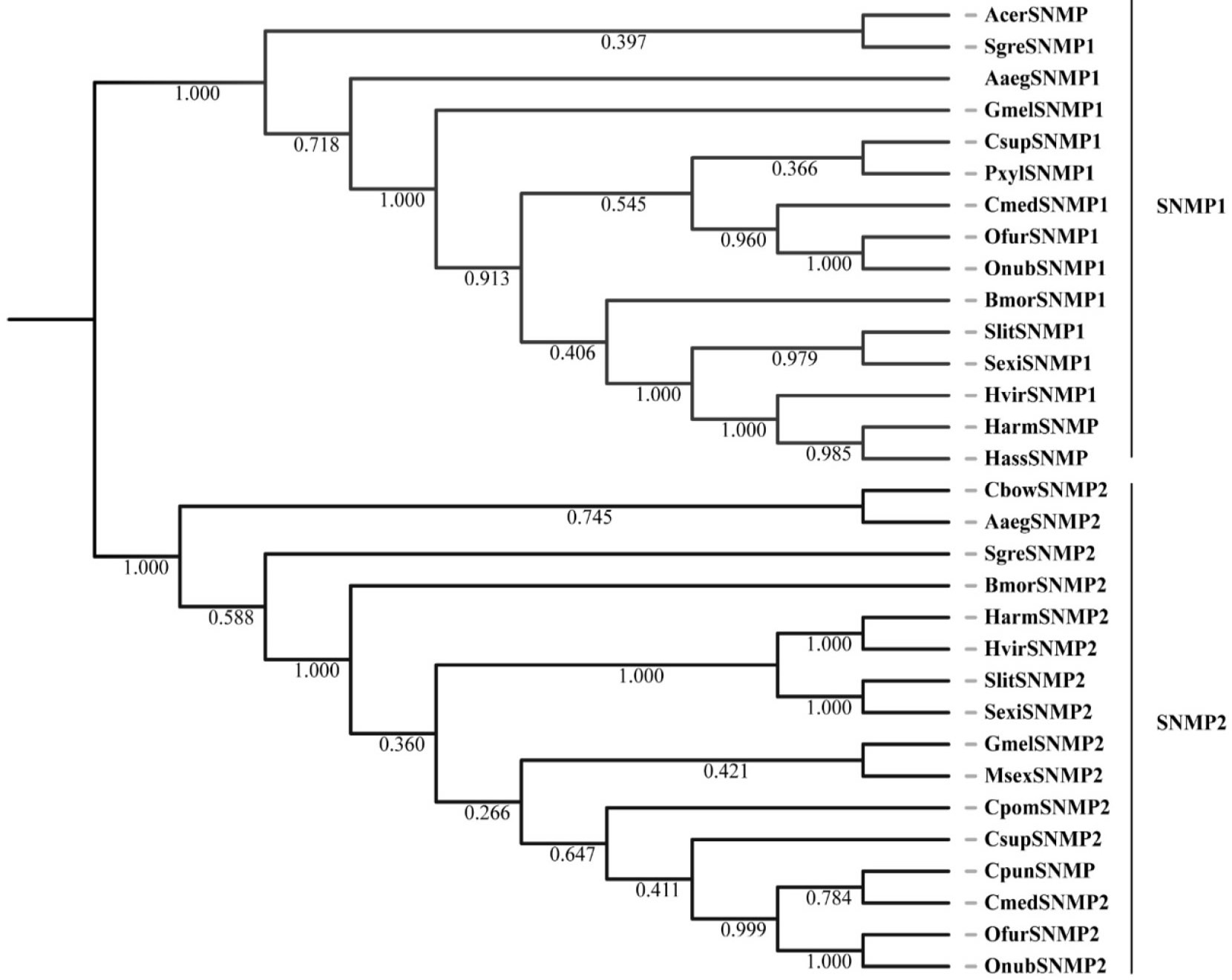
Phylogenetic tree of candidate GmelSNMPs with known sensory neuron membrane protein (SNMP) sequences. Aaeg, Aedes aegypti; Acer, Apis cerana; Bmor, Bombyx mori; Cbow, Colaphellus bowringi; Cmed, Cnaphalocrocis medinalis; Cpom, Cydia pomonella; Cpun, Conogethes punctiferalis; Csup, Chilo suppressalis; Gmel, Galleria mellonella; Harm, Helicoverpa armigera; Hass, Helicoverpa assulta; Hvir, Heliothis virescens; Msex, Manduca sexta; Ofur, Ostrinia furnacalis; Onub, Ostrinia nubilalis; Pxyl, Plutella xylostella; Sexi, Spodoptera exigua; Sgre, Schistocerca gregaria; Slit, Spodoptera litura.
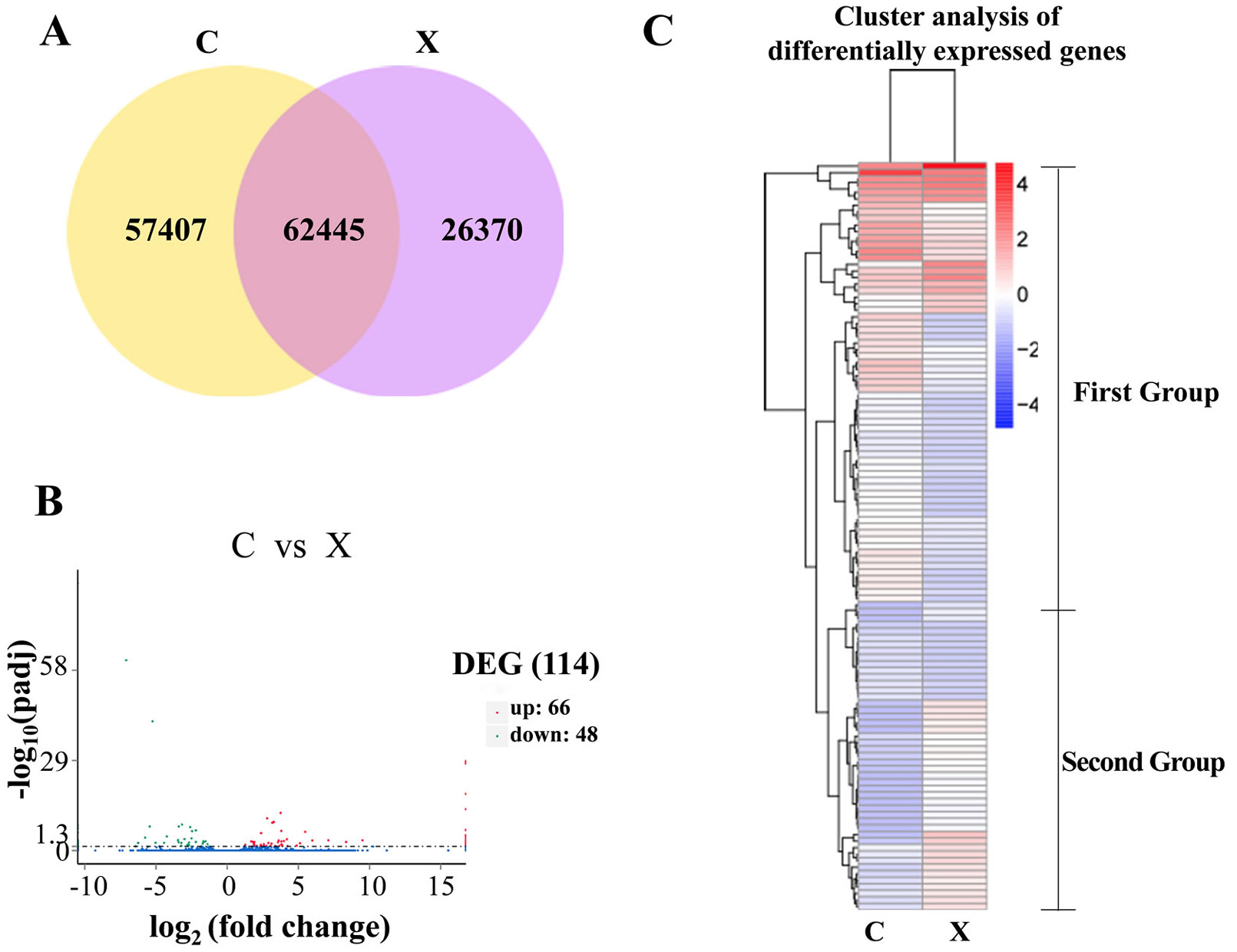
Bioinformatic analyses of differentially expressed genes (DEGs). A, Venn diagrams showing the number of genes expressed in the two groups; B, DEG distribution between the two treatment groups (C, Female; X, Male); C, cluster analysis of DEGs.
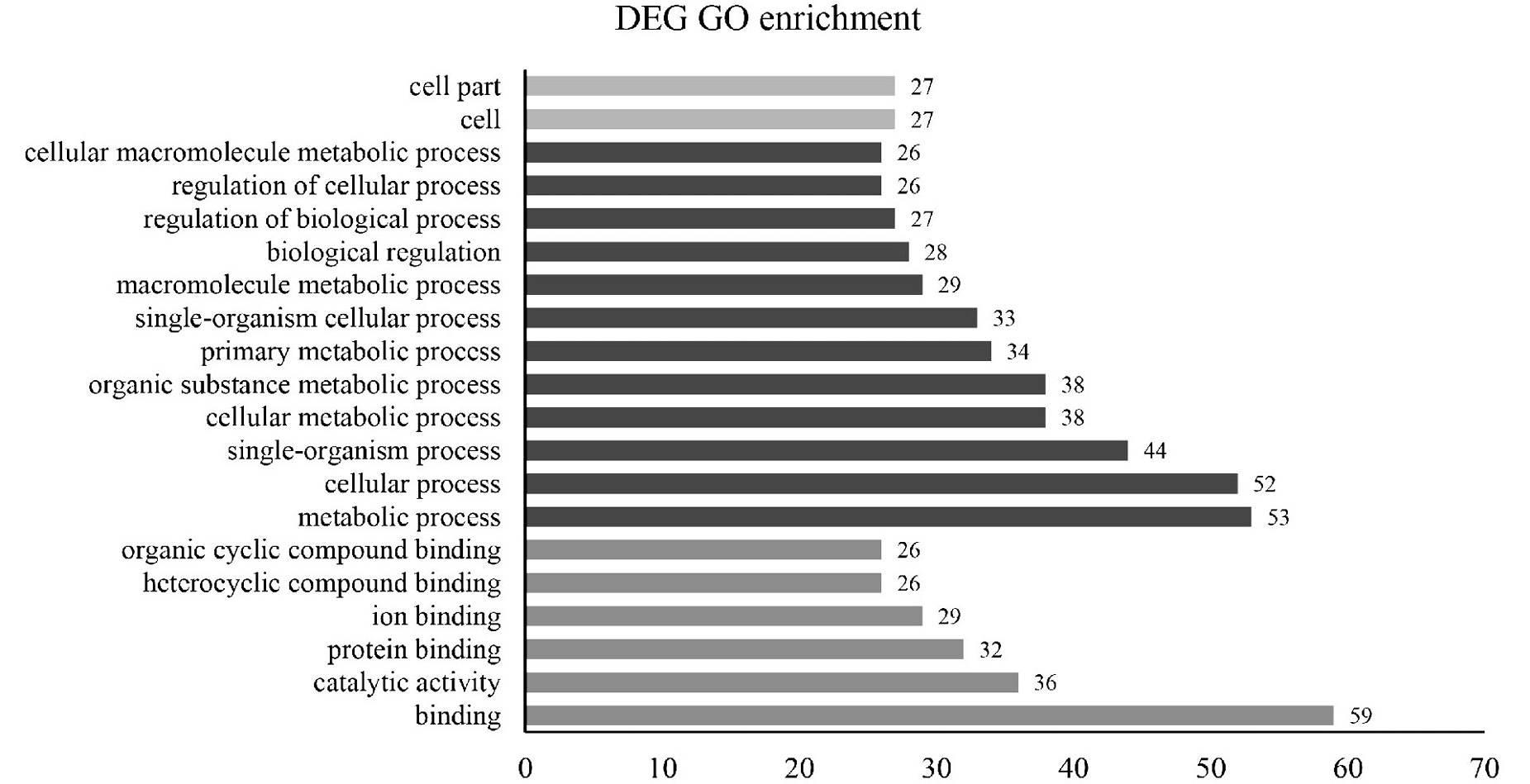
Bar graph showing enriched gene ontology (GO) terms for differentially expressed genes (DEGs) between the female and male antennae.
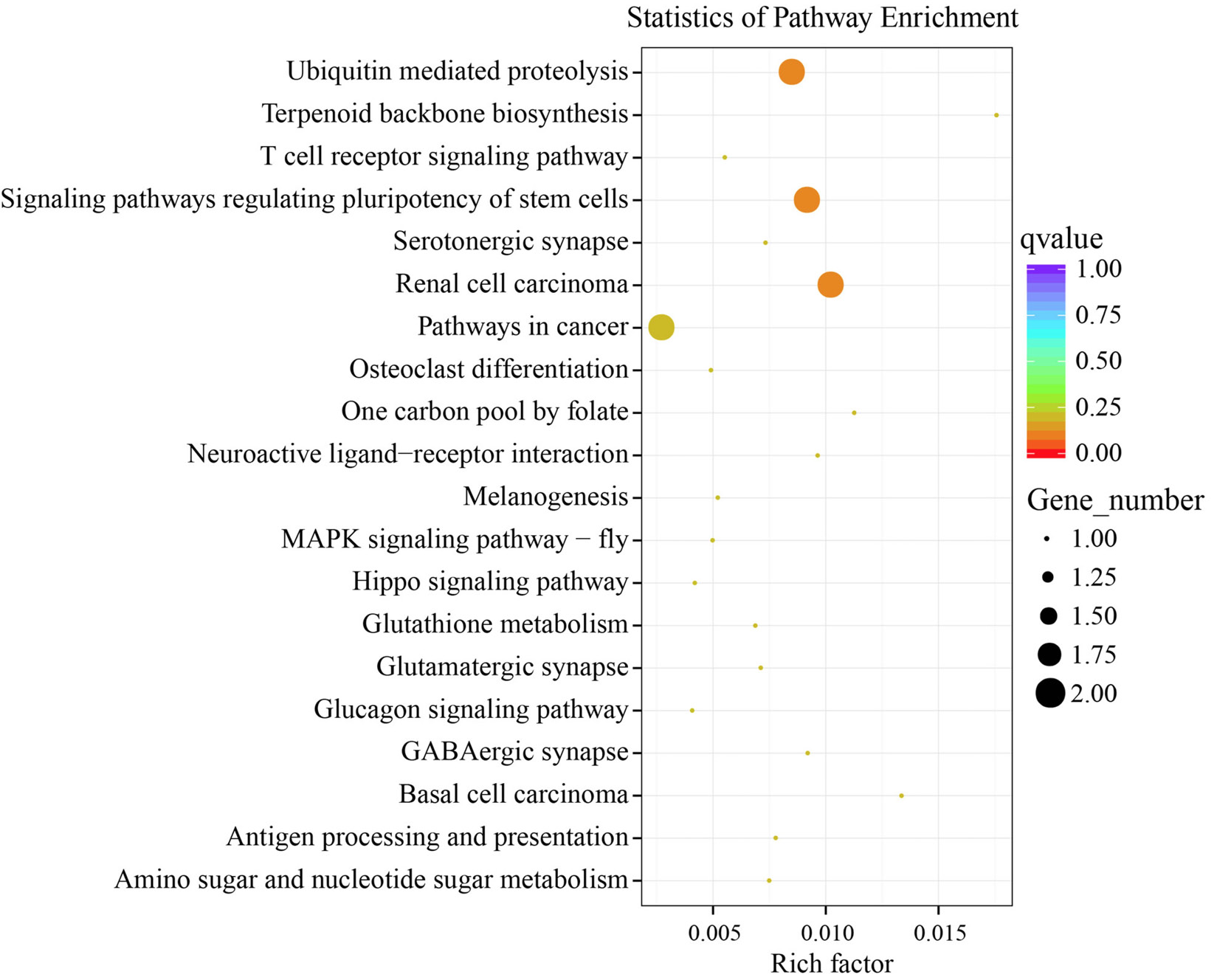
Differentially expressed genes (DEG)-enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.