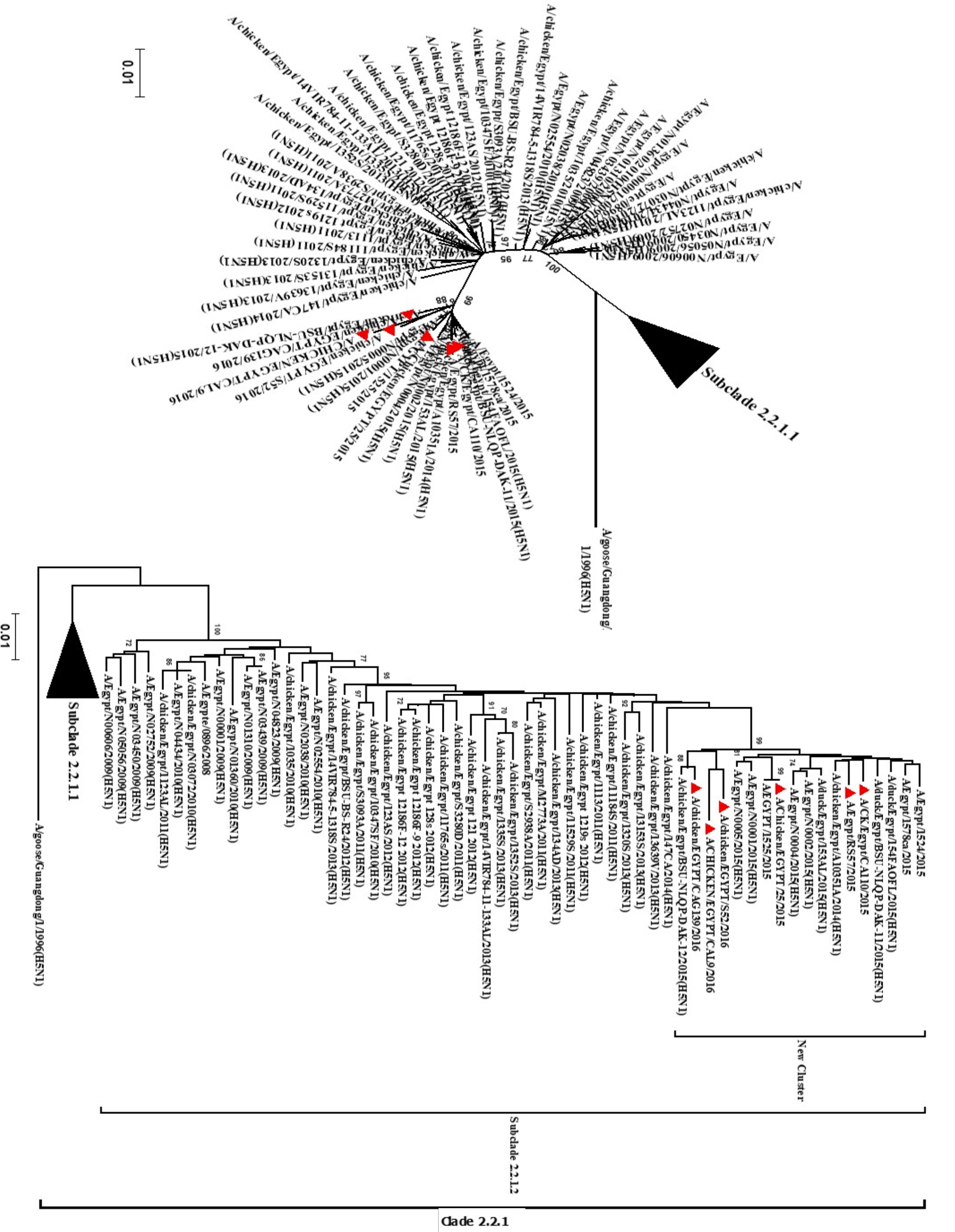
Figure 2:
Phylogenetic tree of haemagglutinin (HA) gene sequences of Egyptian H5N1 viruses showing the continuous evolution of Egyptian H5N1 subclade 2.2.1.2 with the establishment of distinct cluster with close relation to the reported human isolates. The robustness of individual nodes of the tree was assessed using 1000 replications of bootstrap re-sampling of the originally aligned nucleotide sequences. Scale bar represents the number of substitutions per site. The year of isolation and geographical origin of the virus sequences are included in the tree.