Advances in Animal and Veterinary Sciences
Research Article
Isolation, Antigenic and Molecular Characterization of Sheeppox Virus from Clinical Cases in Egypt
Rabab T. Hassanien, Ahmed F. Afify*, Hala K. Abdelmegeed, Nadia M.H. Danial
Virology research department, Animal Health Research Institute, Agriculture Research Center (ARC), 12618 Dokki, Giza, Egypt.
Abstract | Sheep pox is a viral diseases of sheep characterized by fever, generalized papules or nodules, vesicles (rarely), lesions in internal organs and death. Sheeppox virus (SPPV) is the causative agents of Sheeppox, together with goat pox virus (GPV) and lumpy skin disease virus (LSDV) make up the genus Capri poxvirus in the family Poxviridae. The aim of the current study is to monitor and follow up the current situation of Sheeppox (SPPV) in Egypt. A total of thirty-two samples (scabs, biopsies from skin nodules and necropsies from internal organs) were collected from suspected infected sheep with SPPV from two Egyptian governorates (Beheira and Giza) during the summer season of 2018 and 2019. Virus isolation into embryonated chicken eggs (ECEs), transmission electron microscope (TEM), and molecular identification were carried out. Our results revealed that nineteen samples (19/32; 56.4%) were positive based on virus isolation into ECEs. Two representative positive samples were examined using TEM that showed protein filaments projected from the external membrane. Meanwhile, fifteen samples out of nineteen positive samples from virus isolation (15/19; 78.9%) were positive based on polymerase chain reaction (PCR) followed by sequencing and phylogenetic analysis for representative two samples, which revealed high percentage of identity with SPPV reference stains from different countries including Egypt. In conclusions, our findings revealed that the circulation of SPPV within the Egyptian field. Further studies and surveillance are required to monitor the virus evolution and transmission pathways to better understand the virus pathobiology that will help for SPPV control.
Keywords | Capri poxvirus, Sheeppox, Transmission electron microscope, Isolation, Embryonated chicken eggs
Received | October 11, 2020; Accepted | December 08, 2020; Published | January 15, 2021
*Correspondence | Ahmed F. Afify, Virology research department, Animal Health Research Institute, Agriculture Research Center (ARC), 12618 Dokki, Giza, Egypt; Email: [email protected]
Citation | Hassanien RT, Afify AF, Abdelmegeed HK, Danial NMH (2021). Isolation, antigenic and molecular characterization of sheeppox virus from clinical cases in Egypt. Adv. Anim. Vet. Sci. 9(3): 416-421.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.3.416.421
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Hassanien et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Sheeppox virus (SPPV) belongs to the Poxviridae family, genus Capri poxvirus (CaPV). Members of genus CaPV are closely related to each other’s and causing diseases in sheep, goats, and cattle (Tulman et al., 2002). SPPV is highly contagious, notifiable disease, spread directly via aerosols and direct contact with infected animals, and indirectly by contamination of cuts and abrasions (Kitching and Carn, 2004).
Sheeppox virus is endemic in the Middle East (including Egypt), Iran, Afghanistan, North Africa, Turkey, Iraq, India and South-Eastern Europe (OIE, 2017). SPPV causes high economic losses due to high mortality, abortions and loss of market value of the affected animals (OIE, 2017). The morbidity and mortality rates might be different according the animal breed, immune status, infectious strains and virus load. Mortality rate can reach up to 50% in fully susceptible flock and as high as 100% in young animals (OIE, 2017). Moreover, SPPV outbreaks have direct consequences on trade, exportation, and establishment of intensive livestock production (OIE, 2017). There is no specific treatment for Sheeppox infection unless hygienic measures to control the secondary bacterial infection along with cleaning, well ventilation and balanced diet (Senthikumar and Thirunavukkarasu, 2010). Until now, vaccination remains the most effective way to control CaPV infections (Mangana et al., 2008).
Laboratory diagnosis of Poxvirus by electron microscopy (EM) is based on the morphological identification of virus particles in infected tissue samples. EM can be used for differential diagnosis for skin lesions induced by other viruses including SPPV (Yadav et al., 2010). In addition, polymerase chain reaction (PCR) considered a rapid, sensitive, and highly specific approach for detection and differentiation between SPPV and other similar diseases affecting sheep based on the open reading frame 103 (ORF-103) gene for genotyping and phylogenetic analysis (Zhu et al., 2013).
The aim of this study is to isolate, and identify Sheeppox virus (SPPV) based on isolation into embryonated chicken eggs (ECEs), transmission electron microscopy (TEM) and PCR followed by sequencing and phylogeny compared to recent available strains worldwide on public domains (NCBI).
MATERIALS AND METHODS
Ethical approval
International, national, and/or institutional guidelines for the samples collection were followed with the approval of the Local Ethics Committee on Animal Experimentation in Animal Health Research Institute, Agriculture Research Center (ARC); Egypt.
Samples collection and preparation
A total of thirty-two samples including scabs, biopsy from skin scabs and necropsy from internal organs were collected from suspected infected cases with SPPV from two Egyptian governorates; Beheira (21 samples), and Giza (11 samples).
Samples were collected during the summer season of 2018 and 2019. Lesion material was homogenized and prepared following the OIE protocols for virus isolation and molecular detection (OIE, 2017), Briefly, tissues and scabs were homogenized using sterile scissors and forceps, then grinded against sterile sand in a sterile pestle and mortar. An equal volume of sterile phosphate buffered saline (PBS) containing sodium penicillin (1000 IU/ml), streptomycin sulphate (1 mg/ml), mycostatin (100 IU/ml) or fungizone (2.5 μg/ml) and neomycin (200 IU/ml) to 10% final concentration. The homogenized suspension was freeze– thawed three times then clarified by centrifugation at 600 xg for 10 minutes. The supernatant was filtered through a 0.45 μm pore size filter, then aliquoted into sterile 1ml Eppendorf tubes and stored at -80 °C till used.
Virus isolation and transmission electron microscope (TEM)
Specific pathogen free embryonated chicken eggs (SPF ECEs) (12 days old age) were used for virus isolation through inoculation of 100 µl of the prepared samples. The inoculated eggs were incubated at 37°C with 40-60% humidity with candling daily. After five days, the chorio-allantoic membranes (CAMs) were collected and examined for the presence of the characteristic pock lesions of Poxviruses followed by three blind egg passaging were carried out using the collected CAMs.
The collected CAMs with pock lesions were divided into two parts; first part was used in molecular and genetic characterization while the second part was preserved on glutaraldehyde 4% for TEM examination (Mahmoud et al., 2010). TEM was conducted out at EM unit, Central research laboratories, Faculty of Agriculture, Cairo University.
Molecular and genetic characterization of sheeppox virus
DNA was extracted from the prepared CAMs using the QIAamp minielute virus spin kit (Qiagen, Germany, GmbH) according to manufacturing instructions.
ORF 103 gene of Capri pox was amplified as previously described Zhu et al. (2013). The oligonucleotide primers were supplied by Metabion®, Germany, as described in (Table 1).
The PCR products were separated by electrophoresis on 1.5% agarose gel (Applichem, Germany, GmbH) The gel was photographed on a gel documentation system (Alpha Innotech, Biometra) ™ and the data was analyzed through computer software.
The PCR products of two representative samples (2/15) were purified using QIAquick PCR Product extraction kit. (Qiagen, Valencia). Bigdye Terminator V3.1 cycle sequencing kit (Perkin-Elmer)™ was used for sequencing reaction using Applied Biosystems genetic analyzer 3500 (HITACHI, Japan)™. A BLAST® analysis (Basic Local Alignment Search Tool) was initially performed to check the sequence identity (Altschul et al., 1990). The phylogenetic tree was constructed by the MegAlign module of DNASTAR Lasergene version 12.1 using maximum likelihood trees with 1000 bootstrap replicates (Tamura et al., 2013).
RESULTS AND DISCUSSION
CaPVs infection have been listed as an OIE notifiable disease, widely distributed causing high mortality of lambs and immune suppression of both sexes and all ages (OIE, 2017).
In addition, CaPVs have a major impact on the international trade of livestock and their byproducts (Babiuk et al., 2008). With the increasing number of SPPV outbreaks in both sheep and goats with absence of regular vaccination; especially for shepherds in desert areas, we were initiated to carry out an antigenic and molecular characterization of the recent Egyptian SPPV strains to understand its relationship to the previously isolated and published poxviruses. During summer season of 2018 and 2019, samples were collected from suspected clinical cases of sheep based on the clinical signs and disease history. In spite of the disease could be identified based on clinical signs, various laboratory tests have to be carried out for confirmation and differential diagnosis from other diseases which produce similar clinical findings (CPD, bovine herpes virus type-2 and bluetongue virus) (Kottaridi et al., 2006). SPF ECEs were used for the virus isolation by inoculation of the prepared samples on CAM for development of characteristic pock lesions (Grist et al., 1979). Nineteen samples were positive (19/32; 59.3%) based on virus isolation and showed characteristic pock lesions on CAMs (Figure 1). While the infected animals were clinically diagnosed as SPV; it may be due to an inaccurate selection for the sampling site or improper preparation for the samples or may be lesions were caused by other causative agent rather than SPPV (Mahmoud et al., 2010).
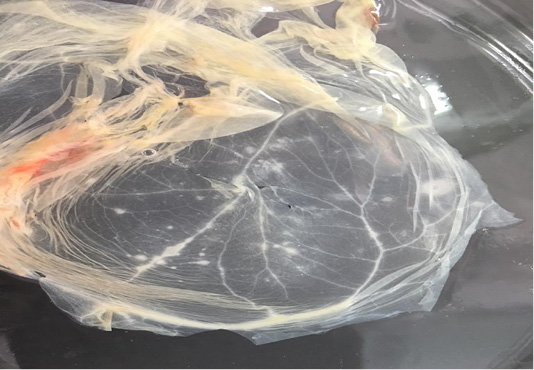
Figure 1: Characteristic pock lesions on CAM of SPF-ECE after three blind successive passages. (Multiple small, opaque and circular).
CAMs were subjected to TEM for confirmation of SPPVs, which verified an approximately 260-320 nm size oval shape viral particles with negative staining with protein filaments projected from the external membrane which is characteristic for CaPVs (Torfason and Gunadottir, 2002; Guo et al., 2003; Mahmoud et al., 2010) as seen in (Figure 2).
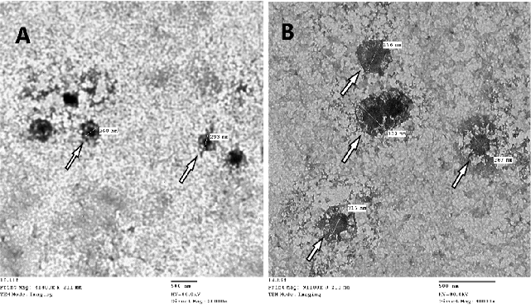
Figure 2: Image of Sheep poxvirus particles visualized by transmission electron microscopy where white arrow points at the virus. SPV particle size ranges from 260-320nm, image A: magnification at X20.000 – B: magnification at X40.000
Although virus isolation is considered the gold standard for SPPV detection and EM is a benchmark method for confirmation, both approaches are time consuming, low sensitivity and need well trained analyst (Lard et al., 1991; Rosenbusch and Reed, 1983; Wittek et al., 1980).
A quite more efficient approach is nucleic acid amplification as PCR, which is one of the most precious tools to identify all infectious diseases of veterinary importance (Mangana-Vougiouka et al., 1999, 2000; Inoshima et al., 2001, 2002; Billinis et al., 2001). Our results showed that SPPV was identified by PCR in fifteen samples out of the nineteen isolated samples on CAMs (15/19; 78.9%). These positive samples gave specific M.W band at 570bp by molecular size (Figure 3). The variation between the isolations results and PCR might be due to low specificity of isolation on ECEs, as many other viruses can be isolated on CAMs (Kottaridi et al., 2006).
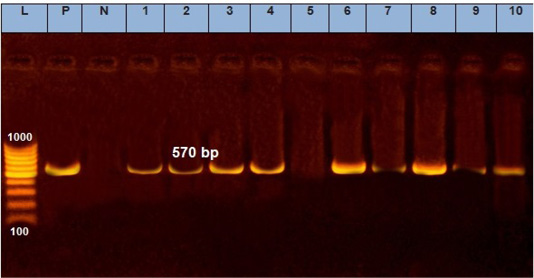
Figure 3: Electrophoresis of the amplified product of PCR (570bp). L: 100 bp DNA marker, p: positive control, N: negative control, from 1 to 10 samples.
Partial nucleotide sequence of ORF 103 gene of our strains named (SPPV_AHRI_1) and (SPPV_AHRI_2) were submitted to the Genbank, obtained the accession numbers MT210507 and MT210508.
Table 1: Primers sequences, target genes and amplicon sizes used in detection of SPV N.A.
| Reference | Amplicon size | Primer sequence (5'-3') | Target gene | Target virus |
| 570 bp | 5'ATGTCTGATAAAAAATTATCTCG 3' | ORF103 | Capri pox | |
| 3'ATCCATACCATCGTCGATAG 5' |

Table 2: Percentage of nucleotides identity between SPPV AHRI strains and SPPV reference strains published on GenBank based on partial sequence of ORF103.
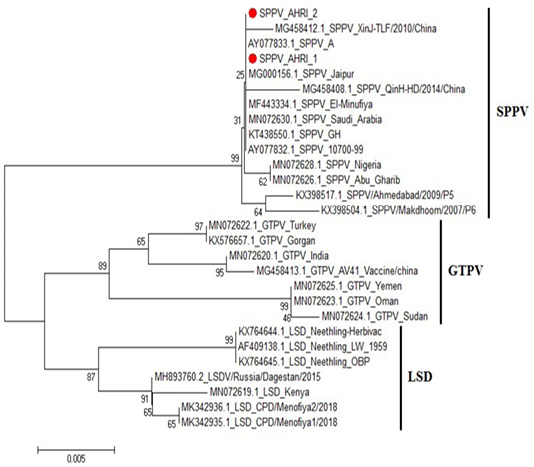
Figure 4: Phylogenetic tree for ORF103 partial nucleotide sequence of sheep poxvirus strain (SPPV_AHRI_1) and (SPPV_AHRI_2) (Acc. No MT210507 and MT210508) (Red circle) compared with other reference strains in GenBank database. The scale bar represents the number of substitutions per nucleotide.
CaPVs have highly conserved genome, with 96% nucleotide identity (Origin and evolution of Poxviruses, 2008). Sequence Analysis of the aligned strains revealed that the two strains have complete identity (at the nucleotide level) with MN072630 (Saudi Arabia 2019), MG000156 (India 1981) and KT438550 (China 2013), AY077832 (USA 2006). Meanwhile, they showed 99.8% identity with MG458412 (China 2010), MN072628 (Nigeria 2019) and MN072626 (Iraq 2019), while 99.6% identity with KX398517 (India 2009) and 99.5% in SPV strain KX398504 (India 2017).
On the other hand, comparing to goat pox viruses (GPVs) the identity was 97.2% with MN072622 (Turkey 2019) and KX576657 (Belgium 2016); 96.7% identity with MN072623 (Oman 2019) and MN072625 (Yemen 2019), and 96.5% identity with MN072624 (Sudan 2019), Figure 4, Table 2. These results agrees with that of (Tulman et al., 2002).
CONCLUSIONS
Sheeppox disease is a growing problem in sheep production causing great economic losses. In this study SPPV was successfully isolated and identified from diseased sheep in 2019, molecular characterization revealed nearly complete identity with all recent SPPVs strains; local and non-neighboring countries which confirms (relatively) stability of SPV genome, that requires further broad, statistically designed and observational analytical epidemiological studies (cohort studies) to systematically evaluate and follow up SPPV prevalence and any potential evolution in circulating strains.
Author’s Contribution
First author conceived of the presented idea and collected samples. Second author developed the theory and performed the computations. All authors carried out the experiment. First and second authors wrote the manuscript with supervision of third and forth authors. All authors discussed the result and contributed to the final manuscript.
Conflict of interest
The authors have declared no conflict of interest.
References






