Epitopes Determination for OMICRON: The COVID-19 New Variant
Epitopes Determination for OMICRON: The COVID-19 New Variant
Saba Rehman1, Faisal Salih Hayat1, Sadia Norin2, Abdul Aziz1, Siddiq Ur Rahman1* and Noor ul Haq1*
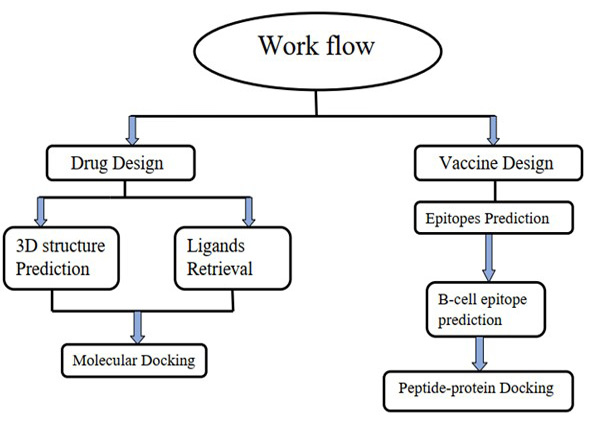
The workflow of drug and vaccine design for COVID-19 omicron variant.
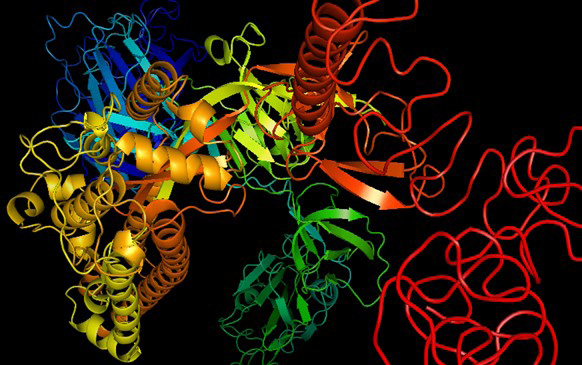
3D structure of COVID-19 variant OMICRON (B.1.1.529). The 3D structure analysis revealed that the structure contains 18 alpha helixes, 13 beta pleated sheets and greater number of the random coils whose theoretical isoelectric point is 7.14.
The protein (omicron variant) docked with ligand molecules. The residues PHE830 and ASP864 show interactions with the ligand REPAGLINIDE (PUBCHEM ID 65981) having bond length of 2.64 while GLY254 and ASP140 having bond length 3.23 was involved in interactions with ligand ENT-NATEGLINIDE (PUBCHEMID 60026).
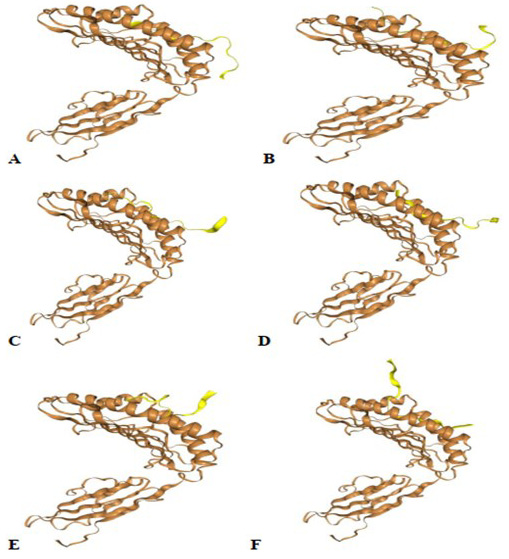
Protein-peptide docking using HPEPDOCK server. (A) GVSVITPGTNTSNQVA, (B) CLIGAEYVNNSYECD, (C) PQIITTDNTFVSGNCD, (D) LRSYSFRPTYGVGHQP, (E) GWTAGAAAYYVGYLQP, (F) AGTITSGWTFGAGAAL