Biological and Molecular Detection of Fig latent trichovirus in Naturally Infected Fig Plants
Biological and Molecular Detection of Fig latent trichovirus in Naturally Infected Fig Plants
Hanaa H.A. Gomaa1*, Dalia Y.Z. Amin1, Mona A. Ismail1 and Khalid A. El-Dougdoug2
Field fig trees showing viral symptom on leaves.
Reaction of host plants inoculated with isolated virus showing none visible symptoms, slightly systemic symptoms on N glutinosa L., N. tabacum cv. white burly, Pepper (Capsucum annum), P. hyprida and tomato, slightly local symptoms on Ch. amaranticolor L. and Ch. quinua.
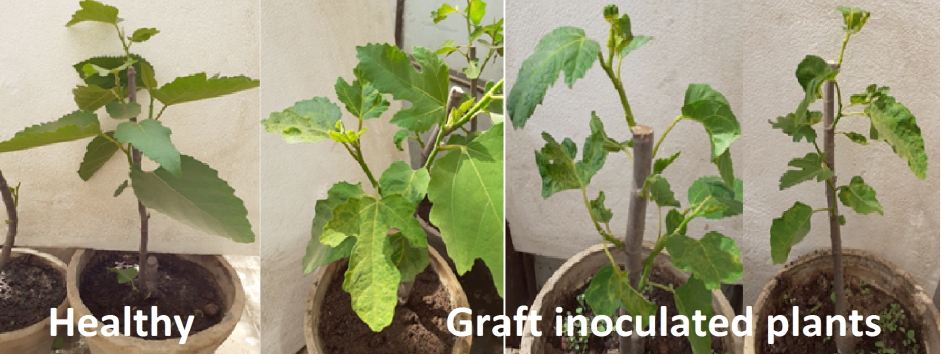
Reaction of fig plants grafting inoculated with isolated virus showing latent systemic symptoms none visible symptoms compared with healthy ones.
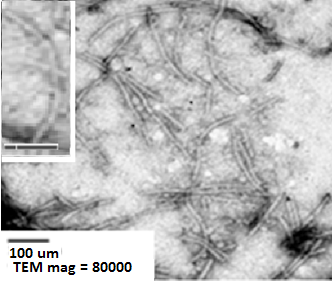
The flexible rod viral particles of FLV-1 detected by negative stain TEM.
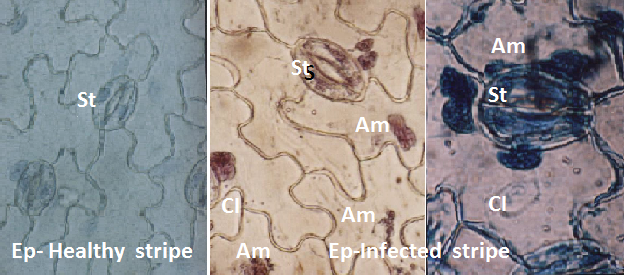
Photograph showing the crystal (Cl) and amorphous (Am) inclusion bodies in cells of epidermal (Ep) strips and stomata (St) of infected and healthy N. glutinosa leaves.
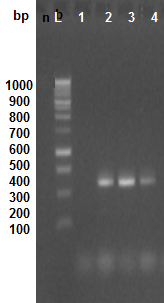
Electrograph of 1.5% agarose gel electrophoresis showing RT-PCR product amplified FLV-CP from extracted RNA from infected fig leaves (1, 2, 3, 4) using (sense CPtr-s and antisense CPtr-s primers . L: DNA ladder weight marker (100 bp ladder).
The nucleotide sequence of partial CP gene (302 bp) of FLV strain (MZ076515).
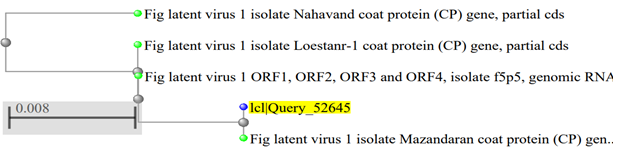
Phylogenetic trees showing the relationship of isolated FLV with Nahavand (KM5167521), I5o5 (FN3775731), Loestanr-1(MG4075531) and Mazandaran, (KM5167521) of fig latent virus isolates recorded in gene bank.
Translation of partial nucleotide sequence of CP gene for FLV strain (MZ076515) produced 100 amino acids.
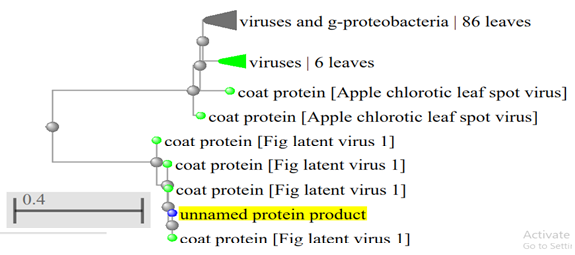
Neighbour joining tree of coat protein gene of FLV-1 CP isolate and 10 isolates published in GenBank based on the amino acid sequence of the CP gene. Numbers represent bootstrap percentage values based on 1000 replicates.
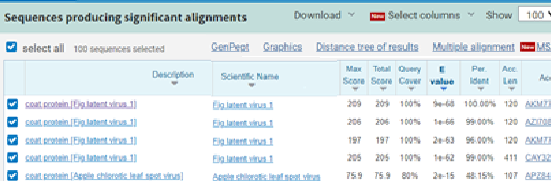
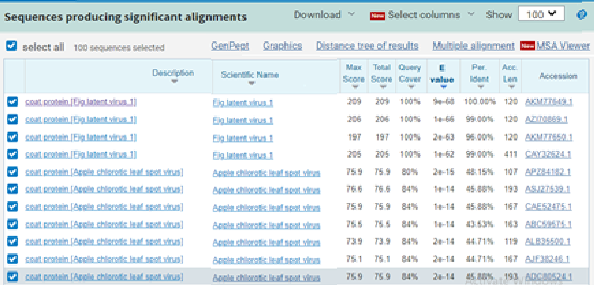
The partial nucleotide sequences producing significant alignments of FLV (MZ076515) CP gene with four FLV isolates recorded in gene bank.
Nucleotide distances and standard error between coat protein gene of FLV isolate and isolates published in GenBank.
Classification and characteristics of Fig latent virus.