Bioinformatics Analysis, Cloning and Expression of Spirometra erinaceieuropaei Fatty Acid-Binding Protein
Lin Huang1, Ling Mai2, Keyan Zhong1 and Xinjun Chen3*
1Experimental Animal Center for Teaching, Hainan Medical University, Haikou 571199, P.R. China.
2Department of Pathology, Danzhou People’s Hospital, Danzhou 571747, P.R. China.
3Laboratory of Pathogenic Biology and Immunology, Hainan Medical University, Haikou 571199, P.R. China.
Lin Huang and Ling Mai contributed equally to this work.
* Corresponding author: chenxj@hainmc.edu.cn
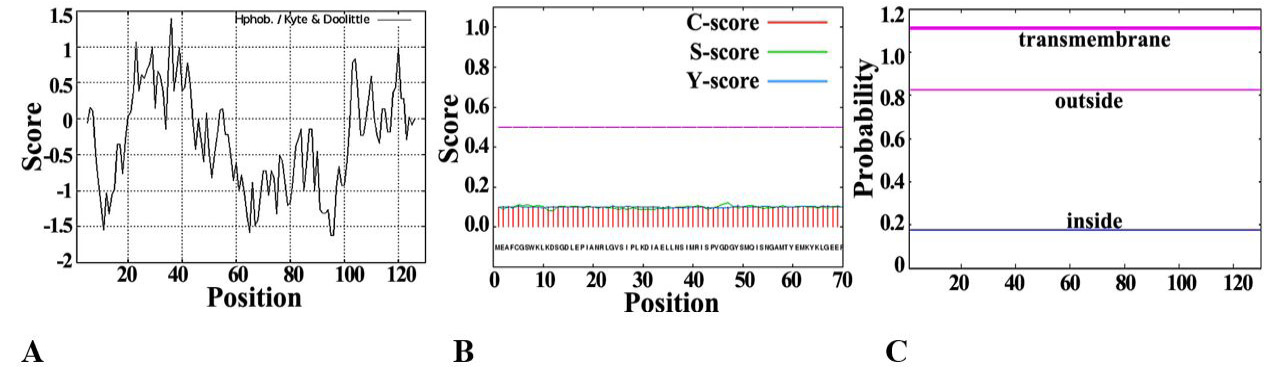
Fig. 1.
Analysis of basic physical and chemical characteristics of SeFABP.
A, Protscale predicts the hydrophilicity/hydrophobicity of SeFABP. B, The signal peptide of SeFABP analyzed by SignalP. C, SeFABP transmembrane domain predicted by TMHMM.
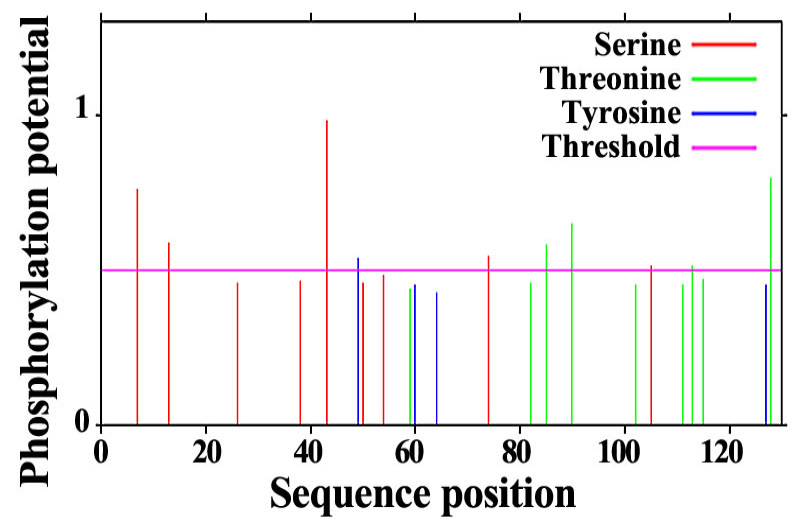
Fig. 2.
The phosphorylation site of SeFABP predicted by NetPho3.1
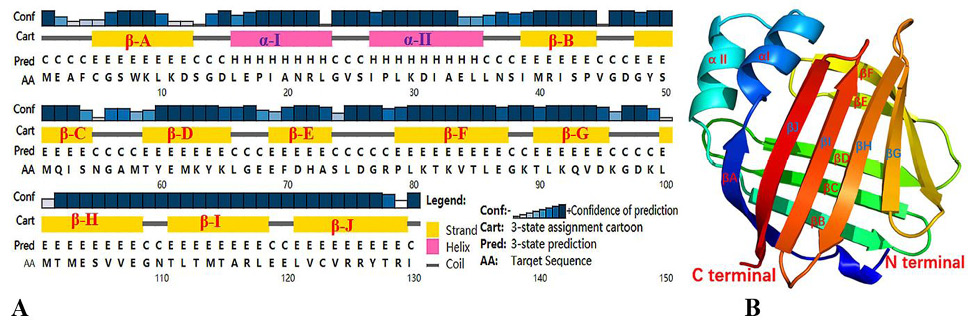
Fig. 3.
Prediction of secondary and tertiary structures of SeFABP. (A) Secondary structure predicted by PSIPRED. (B) 3D structure predicted by Phyre2. Both results showed two short α-helixs, marked α- I, α- II and ten long β-sheets, marked as β-A, β- B... β- J.
Fig. 4.
Quality evaluation of three-dimensional model of SeFABP. (A) The score of Verify3d. (B) Ramachandran diagram analysis of PROCHECK.
Fig. 5.
Multiple sequence alignment of SeFABP and other related parasitic species.
Conservative sites are listed in different colors. All sequences were aligned by Clustal Omega and further edited by Jalview. The letters on consensus represent the same AA residues in these sequences. The percentage similarity of each sequence to SeFABP is displayed in the column marked “ID%”.
Fig. 6.
Phylogenetic analysis of SeFABP with other tapeworms and trematodes.
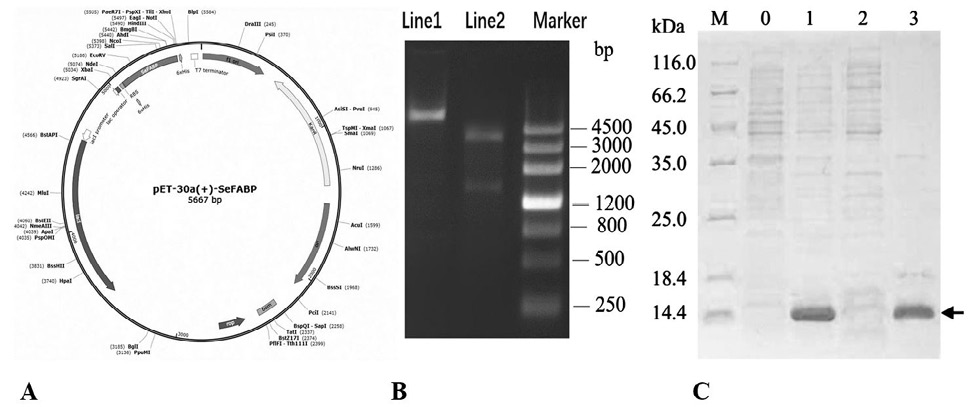
Fig. 7.
The expression and identification of SeFABP.
A, Schematic diagram of expression vector pET-30a(+) -SeFABP. B, The confirmation of the accuracy of vector. Lane 1: plasmid DNA. Lane 2: digested by ApaI and HindIII. C, The expression of SeFABP identified by SDS-PAGE. Lane M, SDS-PAGE protein marker. Lane 0, control (no IPTG). Lane 1, induced at 0.5 mm IPTG at 37 °C for 16 h. Lane 2, supernatant after bacterial fragmentation. Lane 3, sediment after bacterial fragmentation
Fig. 8.
Purification and specificity verification of recombinant SeFABP. A, The purification results of SeFABP in inclusion bodies were analyzed by SDS-PAGE. Lane M, SDS-PAGE protein marker. Lane 1, supernatant after inclusion body dissolution and centrifugation. Lane 2, effluent after incubation of supernatant with Ni IDA. Lane3, elution component of 50 mm imidazole. Lane4-5, elution component of 300 mm imidazole. B, The quality and specificity of FABP were confirmed by SDS-PAGE and Western blot. Lane1, BSA (1.50 μg); Lane 2, SeFABP; M1, SDS-PAGE marker. M2, Western blot marker.