Bioinformatics Analysis Identified the Key Genes of Aspirin and Redox Damaged Yeast Cells
Bioinformatics Analysis Identified the Key Genes of Aspirin and Redox Damaged Yeast Cells
Hui-Ying Chen1,2*, Ping-Chuan Yin1,2, Ya-Nan Lu1,2, Hai-Yun Li1,2*and Yang Shan3
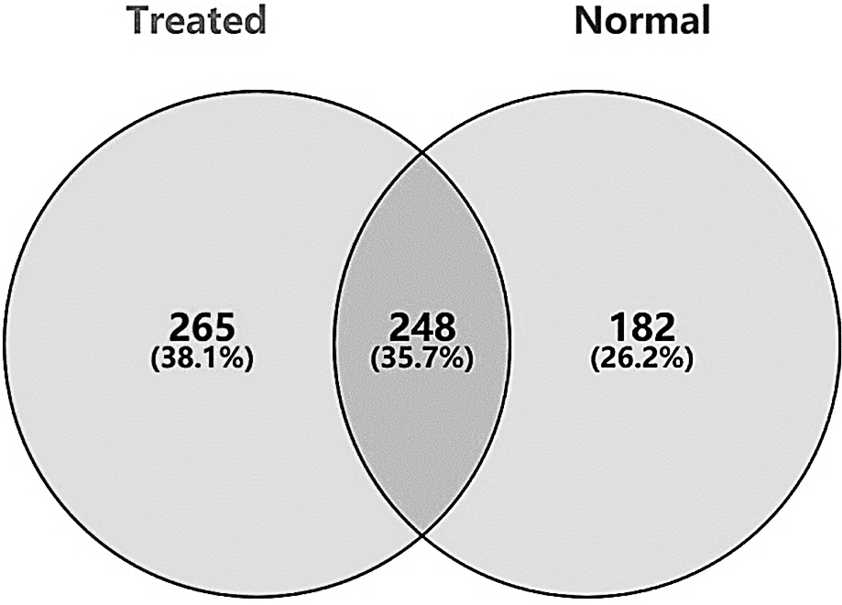
Venn diagram of 248 DEGs from treated and normal.
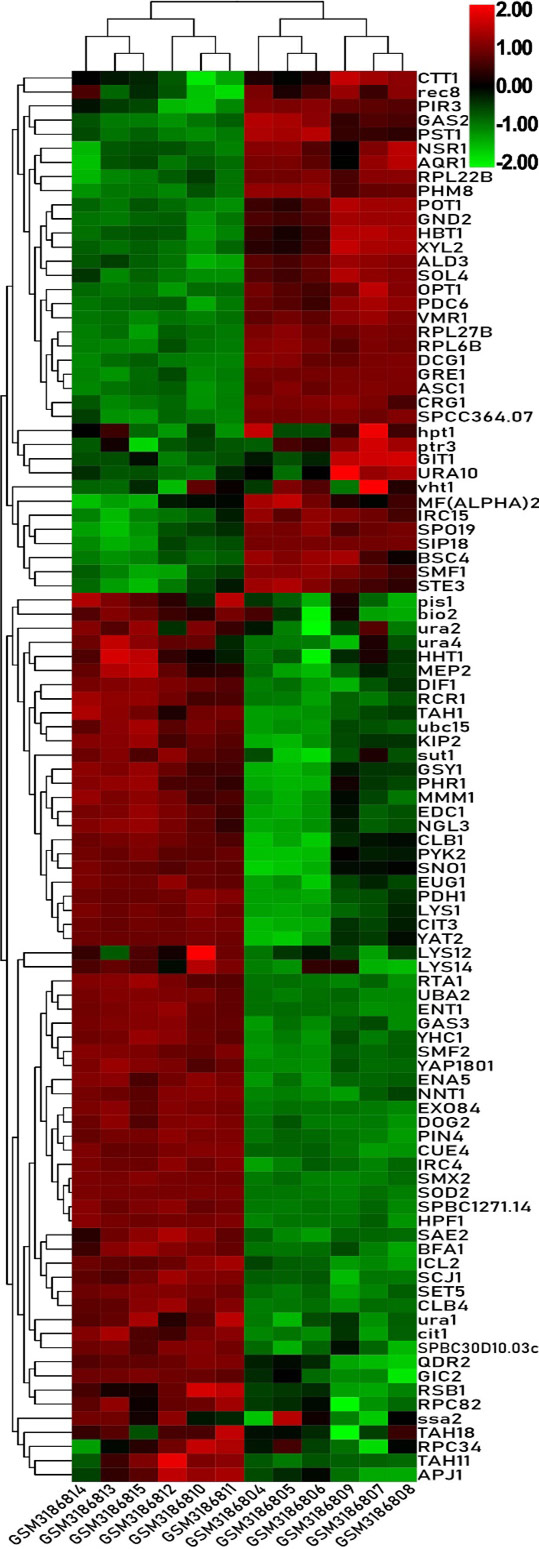
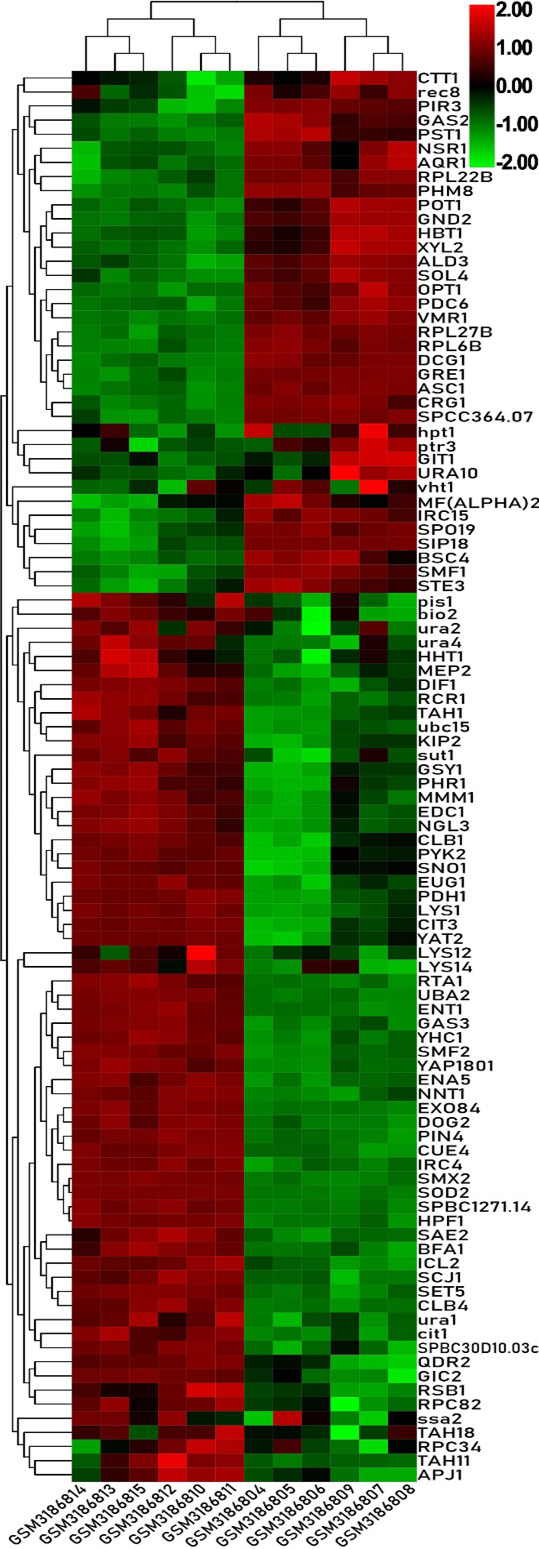
Heat map of the top 100 differentially expressed genes of GSE115660.
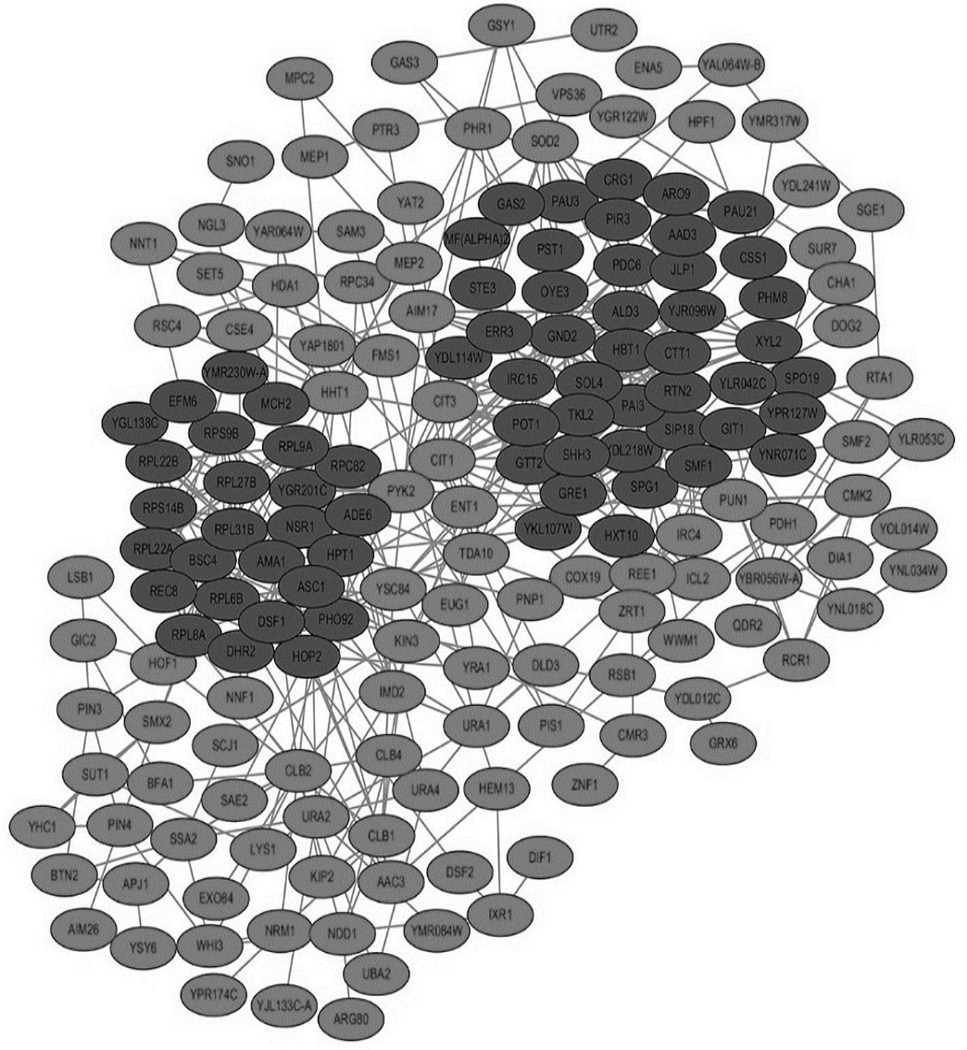
Protein-protein interaction network diagram. Red indicates the up-regulated gene and green indicates the down-regulated gene.
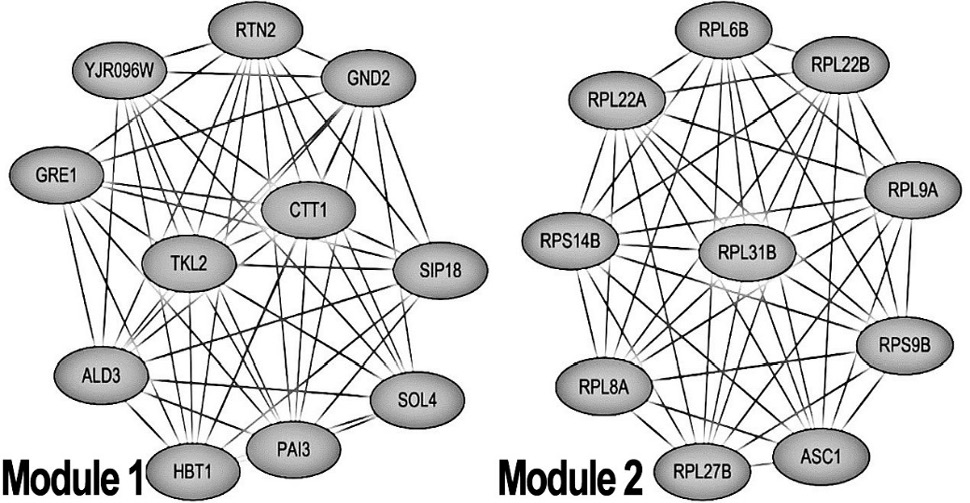
The most significant modules obtained from the PPI network. Module 1 and module 2 are the most important modules in the PPI network identified by the MCODE score > 10.
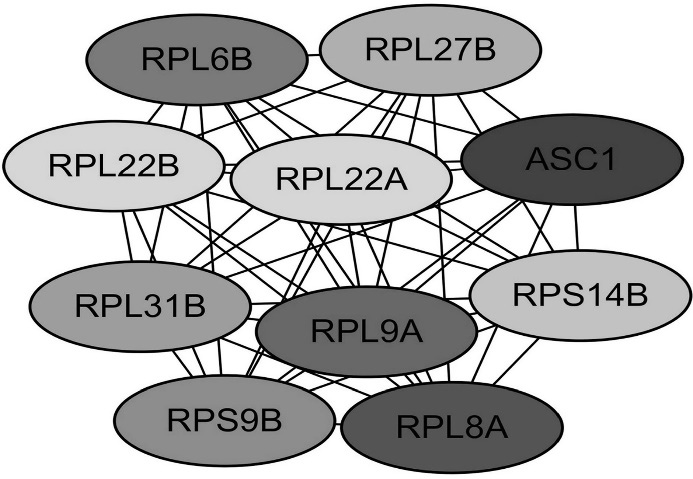
Functional roles of 10 hub genes with MCC ≥10. The darker the color, the higher the score.
Heat map of the top 100 differentially expressed genes of GSE115660.